#include "imgInt.h"#include "fsm.h"#include "heap.h" Include dependency graph for imgLinear.c:
Include dependency graph for imgLinear.c:Go to the source code of this file.
Defines | |
| #define | BACKWARD_REDUCTION_RATE 0.5 |
| #define | FORWARD_REDUCTION_RATE 0.5 |
Functions | |
| static void | ImgLinearUpdateVariableArrayWithId (Relation_t *head, int cindex, int id) |
| static int | ImgLinearQuantifyVariablesFromConjunct (Relation_t *head, Conjunct_t *conjunct, array_t *smoothVarBddArray, int *bModified) |
| static void | ImgLinearConjunctRefine (Relation_t *head, Conjunct_t *conjunct) |
| static void | ImgLinearVariableArrayQuit (Relation_t *head) |
| static void | ImgLinearConjunctQuit (Conjunct_t *conjunct) |
| static Conjunct_t ** | ImgLinearAddConjunctIntoArray (Conjunct_t **array, int *nArray, Conjunct_t *con) |
| static void | ImgLinearFindSameSupportConjuncts (Relation_t *head, int from, int to) |
| static void | ImgLinearClusterSameSupportSet (Relation_t *head) |
| static void | ImgLinearExpandSameSupportSet (Relation_t *head) |
| static int | ImgLinearIsSameConjunct (Relation_t *head, Conjunct_t *con1, Conjunct_t *con2) |
| static bdd_t * | ImgLinearClusteringSmooth (Relation_t *head, Cluster_t *clu, int **failureHistory, int andExistLimit, int bddLimit) |
| static int | ImgLinearClusterUsingHeap (Relation_t *head, double affinityLimit, int andExistLimit, int varLimit, Img_OptimizeType optDir, int rangeFlag, int(*compare_func)(const void *, const void *)) |
| static void | ImgLinearPrintTransitionInfo (Relation_t *head) |
| static void | ImgLinearComputeLifeTime (Relation_t *head, double *paal, double *patl) |
| static void | ImgLinearFreeSmoothArray (array_t *smoothVarBddArray) |
| static void | ImgLinearPrintMatrixFull (Relation_t *head, int matrixIndex) |
| static void | ImgLinearPrintMatrix (Relation_t *head) |
| static void | ImgLinearCAPOReadVariableOrder (Relation_t *head, char *baseName, int includeNS) |
| static void | ImgLinearCAPOInterfaceVariablePl (Relation_t *head, char *baseName, int includeNS) |
| static void | ImgLinearCAPOInterfaceVariableScl (Relation_t *head, char *baseName) |
| static void | ImgLinearCAPOInterfaceVariableNet (Relation_t *head, char *baseName, int includeNS) |
| static void | ImgLinearCAPOInterfaceVariableNodes (Relation_t *head, char *baseName, int includeNS) |
| static void | ImgLinearVariableOrder (Relation_t *head, char *baseName, int includeNS) |
| static void | ImgLinearCAPOReadConjunctOrder (Relation_t *head, char *baseName) |
| static int | ImgLinearCAPORun (char *capoExe, char *baseName, int brief) |
| static void | ImgLinearCAPOInterfaceAux (Relation_t *head, char *baseName) |
| static void | ImgLinearCAPOInterfaceConjunctPl (Relation_t *head, char *baseName) |
| static void | ImgLinearCAPOInterfaceConjunctScl (Relation_t *head, char *baseName) |
| static void | ImgLinearCAPOInterfaceConjunctNet (Relation_t *head, char *baseName) |
| static void | ImgLinearCAPOInterfaceConjunctNodes (Relation_t *head, char *baseName) |
| static void | ImgLinearConjunctionOrder (Relation_t *head, char *baseName, int refineFlag) |
| static void | ImgLinearConjunctOrderMain (Relation_t *head, int bRefineConjunctOrder) |
| static void | ImgLinearPrintDebugInfo (Relation_t *head) |
| static void | ImgLinearPrintVariableProfile (Relation_t *head, char *baseName) |
| static void | ImgLinearInsertClusterCandidate (Relation_t *head, int from, int to, int nDead, int nVar, double affinityLimit) |
| static void | ImgLinearInsertPairClusterCandidate (Relation_t *head, int from, double affinityLimit, int varLimit, int rangeFlag) |
| static void | ImgLinearBuildInitialCandidate (Relation_t *head, double affinityLimit, int varLimit, int rangeFlag, int(*compare_func)(const void *, const void *)) |
| static bdd_t * | ImgLinearClusteringPairSmooth (Relation_t *head, Cluster_t *clu, int **failureHistory, int andExistLimit, int bddLimit) |
| static void | ImgLinearVariableArrayInit (Relation_t *head) |
| static void | ImgLinearSetEffectiveNumberOfStateVariable (Relation_t *head, int *rangeId, int *domainId, int *existStateVariable) |
| static void | ImgLinearVariableLifeQuit (VarLife_t *var) |
| static void | ImgLinearAddNextStateCase (Relation_t *head) |
| static void | ImgLinearAddSingletonCase (Relation_t *head) |
| static int | ImgLinearCompareDeadAffinityLive (const void *c1, const void *c2) |
| static int | ImgLinearCompareDeadLiveAffinity (const void *c1, const void *c2) |
| static int | ImgLinearCompareAffinityDeadLive (const void *c1, const void *c2) |
| static int | ImgLinearCompareLiveAffinityDead (const void *c1, const void *c2) |
| static int | ImgLinearHeapCompareDeadAffinityLive (const void *c1, const void *c2) |
| static int | ImgLinearHeapCompareDeadLiveAffinity (const void *c1, const void *c2) |
| static int | ImgLinearHeapCompareAffinityDeadLive (const void *c1, const void *c2) |
| static int | ImgLinearHeapCompareLiveAffinityDead (const void *c1, const void *c2) |
| static int | ImgLinearCompareVarIndex (const void *c1, const void *c2) |
| static int | ImgLinearCompareVarSize (const void *c1, const void *c2) |
| static int | ImgLinearCompareVarEffFromSmall (const void *c1, const void *c2) |
| static int | ImgLinearCompareVarEffFromLarge (const void *c1, const void *c2) |
| static int | ImgLinearCompareConjunctDummy (const void *c1, const void *c2) |
| static int | ImgLinearCompareConjunctIndex (const void *c1, const void *c2) |
| static int | ImgLinearCompareConjunctSize (const void *c1, const void *c2) |
| static int | ImgLinearCompareConjunctRangeMinusDomain (const void *c1, const void *c2) |
| static int | ImgLinearCompareVarId (const void *c1, const void *c2) |
| static int | ImgCheckRangeTestAndOverapproximate (Relation_t *head) |
| static void | ImgCountOnsetDisjunctiveArray (Relation_t *head) |
| static int | linearCheckRange (const void *tc) |
| static void | ImgLinearConjunctArrayRefine (Relation_t *head) |
| void | ImgLinearConjunctOrderMainCC (Relation_t *head, int refineFlag) |
| int | ImgLinearClustering (Relation_t *head, Img_OptimizeType optDir) |
| int | ImgLinearPropagateConstant (Relation_t *head, int nextStateFlag) |
| int | ImgLinearOptimizeStateVariables (Relation_t *head, Img_OptimizeType optDir) |
| void | ImgLinearExtractNextStateCase (Relation_t *head) |
| void | ImgLinearExtractSingletonCase (Relation_t *head) |
| void | ImgLinearConnectedComponent (Relation_t *head) |
| void | ImgLinearBuildConjunctArrayWithQuotientCC (Relation_t *head) |
| void | ImgLinearFindConnectedComponent (Relation_t *head, Conjunct_t *conjunct, int cc_index) |
| void | ImgLinearAddConjunctIntoClusterArray (Conjunct_t *base, Conjunct_t *con) |
| void | ImgLinearClusterRelationArray (mdd_manager *mgr, ImgFunctionData_t *functionData, array_t *relationArray, Img_DirectionType direction, array_t **clusteredRelationArrayPtr, array_t **arraySmoothVarBddArrayPtr, array_t **optClusteredRelationArrayPtr, array_t **optArraySmoothVarBddArrayPtr, ImgTrmOption_t *option) |
| Relation_t * | ImgLinearRelationInit (mdd_manager *mgr, array_t *relationArray, array_t *domainBddVars, array_t *rangeBddVars, array_t *quantifyBddVars, ImgTrmOption_t *option) |
| int * | ImgLinearGetSupportBddId (mdd_manager *mgr, bdd_t *f, int *nSize) |
| bdd_t ** | ImgLinearExtractRelationArray (Relation_t *head) |
| array_t * | ImgLinearExtractRelationArrayT (Relation_t *head) |
| int | ImgLinearOptimizeAll (Relation_t *head, Img_OptimizeType optDir, int constantNSOpt) |
| void | ImgLinearOptimizeInternalVariables (Relation_t *head) |
| void | ImgLinearRefineRelation (Relation_t *head) |
| void | ImgLinearRelationQuit (Relation_t *head) |
| int | ImgLinearClusteringIteratively (Relation_t *head, double affinityLimit, int andExistLimit, int varLimit, int includeZeroGain, int useFailureHistory, Img_OptimizeType optDir, int(*compare_func)(const void *, const void *)) |
| void | ImgLinearClusteringByConstraints (Relation_t *head, int includeZeroGain, int varLimit, int clusterLimit, int gainLimit, double affinityLimit, int andExistLimit, int bddLimit, int useFailureHistory, int(*compare_func)(const void *, const void *)) |
| array_t * | ImgLinearMakeSmoothVarBdd (Relation_t *head, bdd_t **smoothCubeArr) |
| static int | ImgLinearCompareVarDummyLarge (const void *c1, const void *c2) |
Variables | |
| static char rcsid[] | UNUSED = "$Id: imgLinear.c,v 1.18 2010/04/10 00:37:06 fabio Exp $" |
| static char | linearVarString [] = "TCNI" |
| Cluster_t * | __clu |
Define Documentation
| #define BACKWARD_REDUCTION_RATE 0.5 |
Definition at line 68 of file imgLinear.c.
| #define FORWARD_REDUCTION_RATE 0.5 |
Definition at line 69 of file imgLinear.c.
Function Documentation
| static int ImgCheckRangeTestAndOverapproximate | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Check range of variables]
Description [Check range of variables]
SideEffects []
SeeAlso []
maxIndex = -1; maxSize = 0; for(i=0; i<nVarArray; i++) { var = varArray[i]; if(varType[var->id] == 2) continue; if(var->nSize > maxSize) { maxIndex = i; maxSize = var->nSize; } }
if(maxIndex == -1) return(1);
Definition at line 4898 of file imgLinear.c.
{
Conjunct_t *conjunct;
VarLife_t **varArray, *var;
int nVarArray, i, k, count;
int index;
int *varType;
int allRangeFlag;
bdd_t *relation, *simpleRelation, *varBdd;
allRangeFlag = 1;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
if(conjunct->nRange != conjunct->nSize) {
allRangeFlag = 0;
break;
}
}
if(allRangeFlag == 1) return(1);
varArray = head->varArray;
varType = head->varType;
nVarArray = head->nVarArray;
count = head->nEffDomain + head->nEffQuantify;
if(count == 0) return(1);
varArray = ALLOC(VarLife_t *, head->nVarArray);
memcpy(varArray, head->varArray, sizeof(VarLife_t *)*head->nVarArray);
qsort(varArray, head->nVarArray, sizeof(VarLife_t *), ImgLinearCompareVarSize);
if(count != 0) {
count = (count / 10);
if(count == 0) count = 1;
}
k=0;
index = 0;
while(1) {
if(index >= count) break;
var = varArray[k++];
if(var->stateVar == 2) continue;
index++;
varBdd = bdd_get_variable(head->mgr, var->id);
for(i=0; i<var->nSize; i++) {
conjunct = head->conjunctArray[var->relArr[i]];
relation = conjunct->relation;
simpleRelation = bdd_smooth_with_cube(relation, varBdd);
if(bdd_equal(relation, simpleRelation)) {
bdd_free(simpleRelation);
continue;
}
bdd_free(relation);
conjunct->relation = simpleRelation;
conjunct->bModified = 1;
head->bModified = 1;
head->bRefineVarArray = 1;
ImgLinearConjunctRefine(head, conjunct);
}
}
free(varArray);
ImgLinearRefineRelation(head);
ImgLinearConjunctOrderMainCC(head, 0);
ImgLinearRefineRelation(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
return(0);
}
 Here is the call graph for this function:
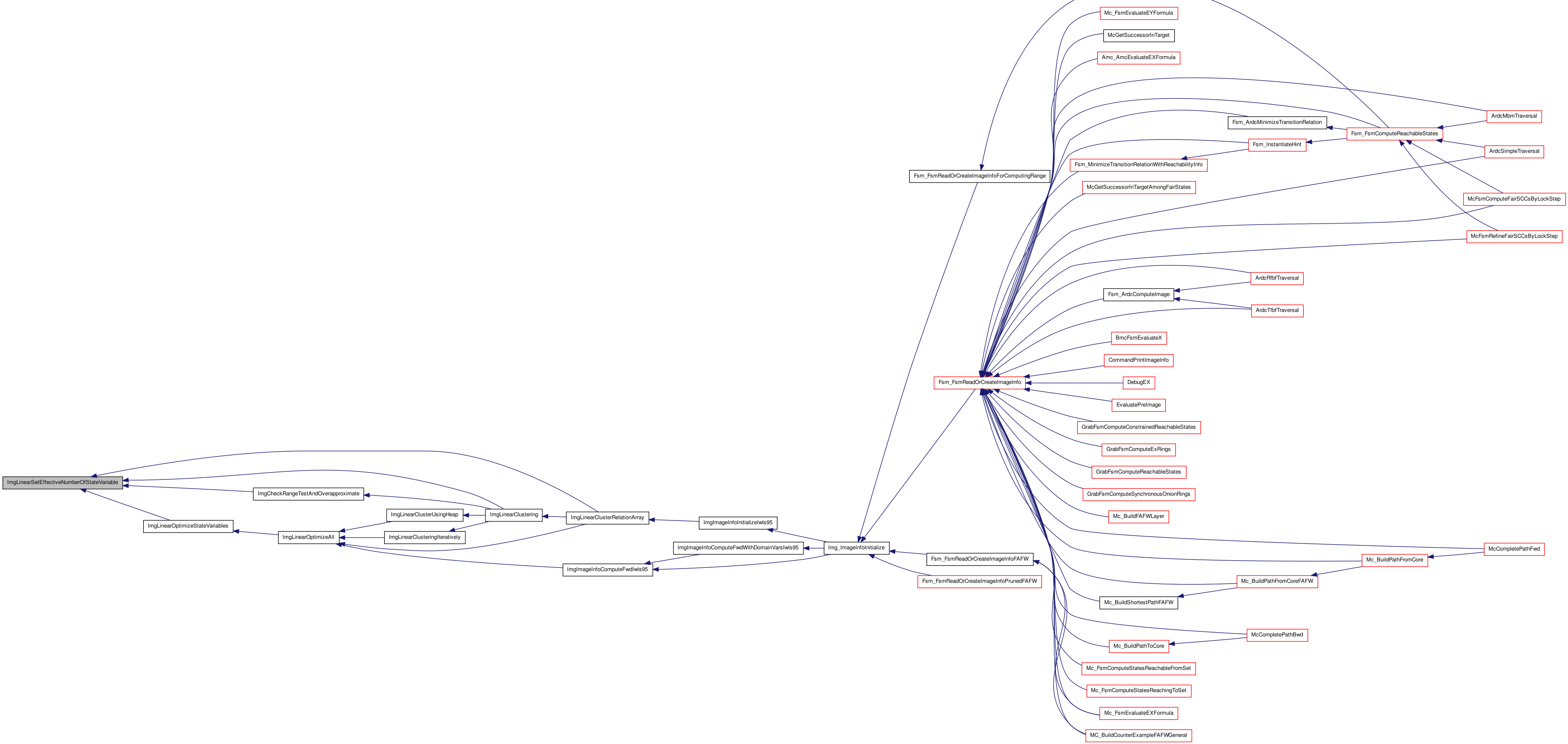
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgCountOnsetDisjunctiveArray | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Count onset of relation array]
Description [Count onset of relation array]
SideEffects []
SeeAlso []
bdd_epd_count_onset(conjunct->relation, bddVarArray, &epd); EpdMultiply2(&tepd, &epd);
Definition at line 4846 of file imgLinear.c.
{
EpDouble tepd;
char strValue[1024];
bdd_t *varBdd;
mdd_manager *mgr;
array_t *bddVarArray;
Conjunct_t *conjunct;
int count, i, j, id;
double onSet;
mgr = head->mgr;
count = 0;
EpdConvert((double)1.0, &tepd);
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
bddVarArray = array_alloc(bdd_t *, 0);
for(j=0; j<conjunct->nSize; j++) {
id = conjunct->supList[j];
varBdd = bdd_get_variable(mgr, id);
array_insert_last(bdd_t *, bddVarArray, varBdd);
}
onSet = bdd_count_onset(conjunct->relation, bddVarArray);
EpdMultiply(&tepd, onSet);
count += conjunct->nSize;
mdd_array_free(bddVarArray);
}
if(count < head->nRange) {
EpdMultiply(&tepd, pow(2, (double)(head->nRange-count)));
}
EpdGetString(&tepd, strValue);
(void) fprintf(vis_stdout, "%-20s%10s\n", "range =", strValue);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static Conjunct_t ** ImgLinearAddConjunctIntoArray | ( | Conjunct_t ** | array, |
| int * | nArray, | ||
| Conjunct_t * | con | ||
| ) | [static] |
Function********************************************************************
Synopsis [Add conjunct into array]
Description [Add conjunct into array]
SideEffects []
SeeAlso []
Definition at line 4582 of file imgLinear.c.
{
if(array)
array = REALLOC(Conjunct_t *, array, (*nArray)+1);
else
array = ALLOC(Conjunct_t *, (*nArray)+1);
array[(*nArray)++] = con;
return(array);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearAddConjunctIntoClusterArray | ( | Conjunct_t * | base, |
| Conjunct_t * | con | ||
| ) |
Function********************************************************************
Synopsis [Add Conjunct into Cluster Array]
Description [Add Conjunct into Cluster Array]
SideEffects []
SeeAlso []
Definition at line 1324 of file imgLinear.c.
{
if(base->clusterArray == 0) {
base->clusterArray = ALLOC(Conjunct_t *, base->nCluster+1);
base->clusterArray[base->nCluster++] = con;
}
else {
base->clusterArray = REALLOC(Conjunct_t *, base->clusterArray, base->nCluster+1);
base->clusterArray[base->nCluster++] = con;
}
base->bClustered = 1;
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearAddNextStateCase | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Add singleton next state variable case into array]
Description [Add singleton next state variable case into array]
SideEffects []
SeeAlso []
Definition at line 4997 of file imgLinear.c.
{
Conjunct_t **newConjunctArray;
int i, j, index;
index = 0;
if(head->nNextStateArray) {
newConjunctArray = ALLOC(Conjunct_t *, head->nSize + head->nNextStateArray);
for(j=0; j<head->nSize; j++)
newConjunctArray[index++] = head->conjunctArray[j];
for(i=0; i<head->nNextStateArray; i++)
newConjunctArray[index++] = head->nextStateArray[i];
head->nSize = index;
head->bModified = 1;
head->bRefineVarArray = 1;
free(head->conjunctArray);
head->conjunctArray = newConjunctArray;
if(head->nextStateArray) free(head->nextStateArray);
head->nextStateArray = 0;
}
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearAddSingletonCase | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Find the case that the relation contains only one next state varaible]
Description [Find the case that the relation contains only one next state varaible]
SideEffects []
SeeAlso []
Definition at line 4319 of file imgLinear.c.
{
Conjunct_t **newConjunctArray;
Conjunct_t **singletonArray;
Conjunct_t *conjunct;
int i, j, index;
int nSingletonArray;
int putFlag;
nSingletonArray = head->nSingletonArray;
singletonArray = head->singletonArray;
if(nSingletonArray == 0) return;
fprintf(stdout, "NOTICE : %d singleton case will be added\n", nSingletonArray);
qsort(singletonArray, nSingletonArray, sizeof(Conjunct_t *), ImgLinearCompareConjunctRangeMinusDomain);
index = 0;
newConjunctArray = ALLOC(Conjunct_t *, head->nSize + nSingletonArray);
putFlag = 0;
for(i=0; i<nSingletonArray; i++) {
conjunct = singletonArray[i];
if(putFlag == 0 && conjunct->nRange > conjunct->nSize-conjunct->nRange) {
putFlag = 1;
for(j=0; j<head->nSize; j++)
newConjunctArray[index++] = head->conjunctArray[j];
}
newConjunctArray[index++] = singletonArray[i];
}
if(putFlag == 0) {
for(j=0; j<head->nSize; j++)
newConjunctArray[index++] = head->conjunctArray[j];
}
head->nSize = index;
head->bModified = 1;
head->bRefineVarArray = 1;
free(head->conjunctArray);
head->conjunctArray = newConjunctArray;
free(singletonArray);
head->singletonArray = 0;
head->nSingletonArray = 0;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearBuildConjunctArrayWithQuotientCC | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Order the each connected component]
Description [The new Relation_t is created for each connected component]
SideEffects []
SeeAlso []
Definition at line 966 of file imgLinear.c.
{
Conjunct_t *conjunct, *tConjunct;
Conjunct_t **connectedArray;
Conjunct_t **conjunctArray;
Conjunct_t **newConjunctArray;
st_table *table;
int i, j, k, l;
int index, id, size;
int *varType, *supList;
st_generator *gen;
char baseName[1024];
if(head->nConnectedComponent == 1) {
size = 0;
newConjunctArray = ALLOC(Conjunct_t *, size+1);
connectedArray = head->connectedComponent[0];
for(j=0; (long)(connectedArray[j])>0; j++) {
tConjunct = connectedArray[j];
newConjunctArray[size++] = tConjunct;
newConjunctArray = REALLOC(Conjunct_t *, newConjunctArray, size+1);
}
free(head->conjunctArray);
head->nSize = size;
head->conjunctArray = newConjunctArray;
head->bModified = 1;
head->bRefineVarArray = 1;
ImgLinearRefineRelation(head);
return;
}
head->includeCS = 1;
head->includeNS = 1;
varType = head->varType;
conjunctArray = ALLOC(Conjunct_t *, head->nConnectedComponent);
for(i=0; i<head->nConnectedComponent; i++) {
connectedArray = head->connectedComponent[i];
table = st_init_table(st_ptrcmp, st_ptrhash);
for(j=0; (long)(connectedArray[j])>0; j++) {
conjunct = connectedArray[j];
size = conjunct->nSize;
supList = conjunct->supList;
for(k=0; k<size; k++) {
id = supList[k];
st_insert(table, (char *)(long)id, (char *)(long)id);
}
for(l=0; l<conjunct->nCluster; l++) {
tConjunct = conjunct->clusterArray[l];
size = tConjunct->nSize;
supList = tConjunct->supList;
for(k=0; k<size; k++) {
id = supList[k];
st_insert(table, (char *)(long)id, (char *)(long)id);
}
}
}
conjunct = ALLOC(Conjunct_t, 1);
memset(conjunct, 0, sizeof(Conjunct_t));
conjunct->index = i;
conjunct->dummy = (long)connectedArray;
supList = ALLOC(int, table->num_entries);
index = 0;
st_foreach_item(table, gen, &id, &id) {
if(varType[id] == 1) conjunct->nDomain++;
else if(varType[id] == 2) conjunct->nRange++;
else if(varType[id] == 3) conjunct->nQuantify++;
supList[index++] = id;
}
conjunct->supList = supList;
conjunct->nSize = index;
conjunctArray[i] = conjunct;
conjunct->index = i;
conjunct->relation = bdd_zero(head->mgr);
}
head->nSize = head->nConnectedComponent;
head->conjunctArray = conjunctArray;
head->bRefineVarArray = 1;
ImgLinearVariableArrayInit(head);
sprintf(baseName, "TopCon");
ImgLinearCAPOInterfaceConjunctNodes(head, baseName);
ImgLinearCAPOInterfaceConjunctNet(head, baseName);
ImgLinearCAPOInterfaceConjunctScl(head, baseName);
ImgLinearCAPOInterfaceConjunctPl(head, baseName);
ImgLinearCAPOInterfaceAux(head, baseName);
ImgLinearCAPORun("MetaPlacer", baseName, 0);
ImgLinearCAPOReadConjunctOrder(head, baseName);
size = 0;
newConjunctArray = ALLOC(Conjunct_t *, size+1);
conjunctArray = head->conjunctArray;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
connectedArray = (Conjunct_t **)conjunct->dummy;
for(j=0; (long)(connectedArray[j])>0; j++) {
tConjunct = connectedArray[j];
newConjunctArray[size++] = tConjunct;
newConjunctArray = REALLOC(Conjunct_t *, newConjunctArray, size+1);
}
ImgLinearConjunctQuit(conjunct);
}
free(conjunctArray);
head->conjunctArray = newConjunctArray;
head->nSize = size;
head->bModified = 1;
head->bRefineVarArray = 1;
ImgLinearRefineRelation(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearBuildInitialCandidate | ( | Relation_t * | head, |
| double | affinityLimit, | ||
| int | varLimit, | ||
| int | rangeFlag, | ||
| int(*)(const void *, const void *) | compare_func | ||
| ) | [static] |
Function********************************************************************
Synopsis [Build data structure for clustering]
Description [Build data structure for clustering]
SideEffects []
SeeAlso []
Definition at line 2108 of file imgLinear.c.
{
int size, i;
head->clusterHeap = Heap_HeapInitCompare(head->nSize, compare_func);
size = head->nSize;
for(i=0; i<size; i++) {
ImgLinearInsertPairClusterCandidate(head, i, affinityLimit, varLimit, rangeFlag);
}
return;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceAux | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface files for CAPO]
Description [Make interface files for CAPO]
SideEffects []
SeeAlso []
Definition at line 2611 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
sprintf(filename, "%s.aux", baseName);
fout = fopen(filename, "w");
fprintf(fout, "RowBasedPlacement : ");
fprintf(fout, "%s.nodes %s.nets %s.pl %s.scl\n",
baseName, baseName, baseName, baseName);
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceConjunctNet | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface files for CAPO]
Description [Make interface files for CAPO]
SideEffects []
SeeAlso []
Definition at line 2492 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
VarLife_t **varArray, *var;
int i, j, nPin;
varArray = head->varArray;
sprintf(filename, "%s.nets", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA nets 1.0\n");
fprintf(fout, "#NumNets : %d\n", head->nVarArray);
qsort(head->varArray, head->nVarArray, sizeof(VarLife_t *), ImgLinearCompareVarId);
nPin = 0;
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(var->nSize == 1) nPin += 2;
else nPin += var->nSize;
}
fprintf(fout, "NumPins : %d\n", nPin);
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(var->nSize == 1)
fprintf(fout, "NetDegree : %d S%d\n", var->nSize+1, var->id);
else
fprintf(fout, "NetDegree : %d S%d\n", var->nSize, var->id);
for(j=0; j<var->nSize; j++)
fprintf(fout, "C%d B\n", var->relArr[j]);
if(var->nSize == 1)
fprintf(fout, "C%d B\n", var->relArr[0]);
}
fclose(fout);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceConjunctNodes | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface files for CAPO]
Description [Make interface files for CAPO]
SideEffects []
SeeAlso []
Definition at line 2440 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
int i;
sprintf(filename, "%s.nodes", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA nodes 1.0\n");
if(head->includeNS) {
if(head->includeCS) {
fprintf(fout, "NumNodes : %d\n", head->nSize+2);
fprintf(fout, "NumTerminals : 2\n");
}
else {
fprintf(fout, "NumNodes : %d\n", head->nSize+1);
fprintf(fout, "NumTerminals : 1\n");
}
}
else if(head->includeCS) {
fprintf(fout, "NumNodes : %d\n", head->nSize+1);
fprintf(fout, "NumTerminals : 1\n");
}
else {
fprintf(fout, "NumNodes : %d\n", head->nSize);
fprintf(fout, "NumTerminals : 0\n");
}
for(i=0; i<head->nSize; i++)
fprintf(fout, "C%d 1 1\n", i);
if(head->includeCS)
fprintf(fout, "C%d 1 1 terminal\n", MAXINT-1);
if(head->includeNS)
fprintf(fout, "C%d 1 1 terminal\n", MAXINT);
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceConjunctPl | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface files for CAPO]
Description [Make interface files for CAPO]
SideEffects []
SeeAlso []
Definition at line 2577 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
int i;
sprintf(filename, "%s.pl", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA pl 1.0\n");
for(i=0; i<head->nSize; i++)
fprintf(fout, "C%d %d 1\n", i, (i+2)*2+1);
if(head->includeCS)
fprintf(fout, "C%d 0 1 / N / FIXED\n", MAXINT-1);
if(head->includeNS)
fprintf(fout, "C%d %d 1 / N / FIXED\n", MAXINT, (head->nSize+2)*2+1);
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceConjunctScl | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface files for CAPO]
Description [Make interface files for CAPO]
SideEffects []
SeeAlso []
Definition at line 2543 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
sprintf(filename, "%s.scl", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA scl 1.0\n");
fprintf(fout, "Numrows : 1\n");
fprintf(fout, "CoreRow Horizontal\n");
fprintf(fout, " Coordinate : 1\n");
fprintf(fout, " Height : 1\n");
fprintf(fout, " Sitewidth : 1\n");
fprintf(fout, " Sitespacing : 1\n");
fprintf(fout, " Siteorient : N\n");
fprintf(fout, " Sitesymmetry : Y\n");
fprintf(fout, " SubrowOrigin : 0\n");
fprintf(fout, " Numsites : %d\n", head->nSize*2+2);
fprintf(fout, "End\n");
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceVariableNet | ( | Relation_t * | head, |
| char * | baseName, | ||
| int | includeNS | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface for CAPO]
Description [Make interface for CAPO]
SideEffects []
SeeAlso []
Definition at line 2828 of file imgLinear.c.
{
int nPin, i, j;
int id, size;
Conjunct_t *conjunct;
bdd_t *relation;
int *supList, *varType;
char filename[1024];
FILE *fout;
nPin = 0;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
relation = conjunct->relation;
if(relation == (mdd_t *)(MAXINT-1)) continue;
supList = conjunct->supList;
if(includeNS) {
if(conjunct->nSize == 1) nPin += 2;
else nPin += conjunct->nSize-conjunct->nRange;
}
else {
if(conjunct->nSize-conjunct->nRange == 1) nPin += 2;
else nPin += conjunct->nSize-conjunct->nRange;
}
}
nPin += head->nDomain;
if(includeNS)
nPin += head->nRange;
varType = head->varType;
sprintf(filename, "%s.nets", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA nets 1.0\n");
fprintf(fout, "#NumNets : %d\n", head->nSize);
fprintf(fout, "NumPins : %d\n", nPin);
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
relation = conjunct->relation;
if(relation == (mdd_t *)(MAXINT-1)) continue;
supList = conjunct->supList;
if(includeNS) size = conjunct->nSize;
else size = conjunct->nSize - conjunct->nRange;
if(size == 1) fprintf(fout, "NetDegree : %d \n", 2);
else fprintf(fout, "NetDegree : %d \n", size);
id = 0;
if(includeNS) {
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
fprintf(fout, "C%d B\n", id);
}
}
else {
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
if(varType[id] == 2) continue;
fprintf(fout, "C%d B\n", id);
}
}
if(size == 1)
fprintf(fout, "C%d B\n", id);
}
fprintf(fout, "NetDegree : %d \n", head->nDomain);
for(i=0; i<head->nVar; i++) {
if(varType[i] == 1)
fprintf(fout, "C%d B\n", i);
}
if(includeNS) {
fprintf(fout, "NetDegree : %d \n", head->nRange);
for(i=0; i<head->nVar; i++) {
if(varType[i] == 2)
fprintf(fout, "C%d B\n", i);
}
}
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearCAPOInterfaceVariableNodes | ( | Relation_t * | head, |
| char * | baseName, | ||
| int | includeNS | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface for CAPO]
Description [Make interface for CAPO]
SideEffects []
SeeAlso []
Definition at line 2750 of file imgLinear.c.
{
int i, size;
VarLife_t **varArray, *var;
int *varType;
char filename[1024];
FILE *fout;
int numUnused;
varArray = head->varArray;
varType = head->varType;
sprintf(filename, "%s.nodes", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA nodes 1.0\n");
size = 0;
if(includeNS) size = head->nVarArray;
else {
for(i=0; i<head->nVarArray; i++) {
if(varType[varArray[i]->id] == 2) continue;
size++;
}
}
numUnused = 0;
for(i=0; i<head->nVar; i++) {
if(varType[i] != 1) continue; /* if it is not domain variable **/
var = head->varArrayMap[i] >=0 ? head->varArray[head->varArrayMap[i]] : 0;
if(var) continue;
numUnused++;
}
if(includeNS) {
for(i=0; i<head->nVar; i++) {
if(varType[i] != 2) continue; /* if it is not range variable **/
var = head->varArrayMap[i] >=0 ? head->varArray[head->varArrayMap[i]] : 0;
if(var) continue;
numUnused++;
}
}
fprintf(fout, "NumNodes : %d\n", size+numUnused);
fprintf(fout, "NumTerminals : 0\n");
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(includeNS == 0 && varType[var->id] == 2) continue;
fprintf(fout, "C%d 1 1\n", var->id);
}
for(i=0; i<head->nVar; i++) {
if(varType[i] != 1) continue; /* if it is not domain variable **/
var = head->varArrayMap[i] >=0 ? head->varArray[head->varArrayMap[i]] : 0;
if(var) continue;
fprintf(fout, "C%d 1 1\n", i);
}
if(includeNS) {
for(i=0; i<head->nVar; i++) {
if(varType[i] != 2) continue; /* if it is not range variable **/
var = head->varArrayMap[i] >=0 ? head->varArray[head->varArrayMap[i]] : 0;
if(var) continue;
fprintf(fout, "C%d 1 1\n", i);
}
}
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceVariablePl | ( | Relation_t * | head, |
| char * | baseName, | ||
| int | includeNS | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface for CAPO]
Description [Make interface for CAPO]
SideEffects []
SeeAlso []
Definition at line 2956 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
VarLife_t *var, **varArray;
int i, index, *varType;
varType = head->varType;
sprintf(filename, "%s.pl", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA pl 1.0\n");
varArray = head->varArray;
for(index=0, i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(includeNS == 0 && varType[var->id] == 2) continue;
fprintf(fout, "C%d %d 1\n", var->id, (index+1)*2+1);
index++;
}
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOInterfaceVariableScl | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface for CAPO]
Description [Make interface for CAPO]
SideEffects []
SeeAlso []
Definition at line 2921 of file imgLinear.c.
{
char filename[1024];
FILE *fout;
sprintf(filename, "%s.scl", baseName);
fout = fopen(filename, "w");
fprintf(fout, "UCLA scl 1.0\n");
fprintf(fout, "Numrows : 1\n");
fprintf(fout, "CoreRow Horizontal\n");
fprintf(fout, " Coordinate : 1\n");
fprintf(fout, " Height : 1\n");
fprintf(fout, " Sitewidth : 1\n");
fprintf(fout, " Sitespacing : 1\n");
fprintf(fout, " Siteorient : N\n");
fprintf(fout, " Sitesymmetry : Y\n");
fprintf(fout, " SubrowOrigin : 0\n");
fprintf(fout, " Numsites : %d\n", head->nVarArray*2+2);
fprintf(fout, "End\n");
fclose(fout);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOReadConjunctOrder | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Read result of CAPO]
Description [Read result of CAPO]
SideEffects []
SeeAlso []
Definition at line 2681 of file imgLinear.c.
{
char ordFile[1024];
char line[1024];
int index, id;
char *next;
FILE *fin;
Conjunct_t *conjunct;
sprintf(ordFile, "%s.ord", baseName);
if(!(fin = fopen(ordFile, "r"))) {
fprintf(stdout, "Can't open order file %s\n", ordFile);
exit(0);
}
index = 0;
while(fgets(line, 1024, fin)){
next = strchr(line, 'C');
next++;
sscanf(next, "%d", &id);
if(id == MAXINT) continue;
if(id == MAXINT-1) continue;
conjunct = head->conjunctArray[id];
conjunct->index = index;
index++;
}
fclose(fin);
qsort(head->conjunctArray, head->nSize, sizeof(Conjunct_t *), ImgLinearCompareConjunctIndex);
head->bModified = 1;
head->bRefineVarArray = 1;
ImgLinearRefineRelation(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearCAPOReadVariableOrder | ( | Relation_t * | head, |
| char * | baseName, | ||
| int | includeNS | ||
| ) | [static] |
Function********************************************************************
Synopsis [Read result of CAPO]
Description [Read result of CAPO]
SideEffects []
SeeAlso []
Definition at line 2991 of file imgLinear.c.
{
char ordFile[1024], line[1024];
FILE *fin;
char *next;
int i, id, index, *permutation, *exist;
sprintf(ordFile, "%s.ord", baseName);
if(!(fin = fopen(ordFile, "r"))) {
fprintf(stdout, "Can't open order file %s\n", ordFile);
exit(0);
}
permutation = ALLOC(int, head->nVar);
exist = ALLOC(int, head->nVar);
memset(exist, 0, sizeof(int)*head->nVar);
index = 0;
while(fgets(line, 1024, fin)){
next = strchr(line, 'C');
next++;
sscanf(next, "%d", &id);
permutation[index++] = id;
exist[id] = 1;
if(includeNS == 0 && head->varType[id] == 1) {
permutation[index++] = head->domain2range[id];
exist[head->domain2range[id]] = 1;
}
}
fclose(fin);
for(i=0; i<head->nVar; i++)
if(exist[i] == 0)
permutation[index++] = i;
free(exist);
bdd_shuffle_heap(head->mgr, permutation);
free(permutation);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCAPORun | ( | char * | capoExe, |
| char * | baseName, | ||
| int | brief | ||
| ) | [static] |
Function********************************************************************
Synopsis [Run batch job of CAPO]
Description [Run batch job of CAPO]
SideEffects []
SeeAlso []
Definition at line 2636 of file imgLinear.c.
{
char logFile[1024];
char capoOption[1024];
char command[1024];
char cpCommand[1024];
FILE *fout;
int cmdStatus;
fout = fopen("seeds.in", "w");
fprintf(fout, "0\n");
fprintf(fout, "460427264\n");
fclose(fout);
if(brief)
sprintf(capoOption, "-replaceSmallBlocks Never -noRowIroning -save -saveXorder");
else
sprintf(capoOption, "-save -saveXorder -clust CutOpt");
sprintf(logFile, "%s.log", baseName);
sprintf(command, "%s -f %s.aux %s > %s",
capoExe, baseName, capoOption, logFile);
cmdStatus = system(command);
sprintf(cpCommand, "cp left2right.ord %s.ord", baseName);
cmdStatus |= system(cpCommand);
unlink("seeds.out");
unlink("left2right.ord");
return (cmdStatus == 0);
}
 Here is the caller graph for this function:
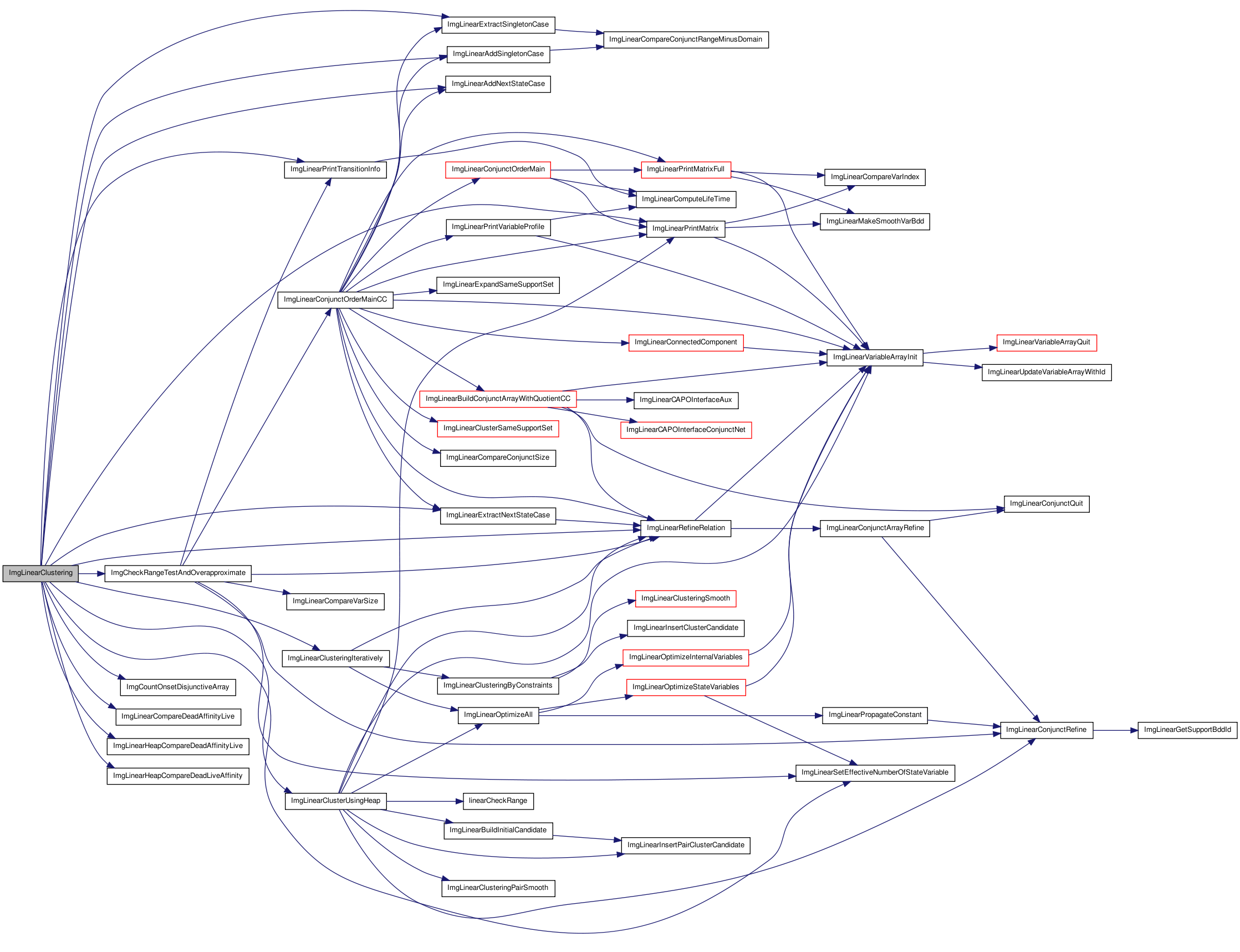
Here is the caller graph for this function:| int ImgLinearClustering | ( | Relation_t * | head, |
| Img_OptimizeType | optDir | ||
| ) |
Function********************************************************************
Synopsis [Main function of clustering]
Description [Main function of clustering]
SideEffects []
SeeAlso []
Definition at line 4678 of file imgLinear.c.
{
int andExistLimit;
int useFailureHistory;
int includeZeroGain;
double affinityLimit;
int bOptimize;
int varLimit;
bOptimize = 0;
affinityLimit = 0.99;
andExistLimit = 10;
varLimit = 200;
fprintf(stdout, "Heap DAL affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
bOptimize |= ImgLinearClusterUsingHeap(head, affinityLimit, andExistLimit,
varLimit, optDir, 0,
ImgLinearHeapCompareDeadAffinityLive);
ImgLinearRefineRelation(head);
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
includeZeroGain = 0;
affinityLimit = 0.3;
andExistLimit = 5000;
varLimit = 50;
useFailureHistory = 0;
if(head->computeRange) {
varLimit = 50;
affinityLimit = 0.8;
andExistLimit = 5000;
fprintf(stdout, "Heap DAL affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
bOptimize |= ImgLinearClusterUsingHeap(head, affinityLimit, andExistLimit,
varLimit, optDir, 0,
ImgLinearHeapCompareDeadAffinityLive);
ImgLinearRefineRelation(head);
ImgLinearExtractNextStateCase(head);
ImgLinearExtractSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddNextStateCase(head);
ImgLinearRefineRelation(head);
}
else
{
fprintf(stdout, "Gain DAL affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
bOptimize |= ImgLinearClusteringIteratively(head, affinityLimit,
andExistLimit, varLimit,
includeZeroGain, useFailureHistory, optDir,
ImgLinearCompareDeadAffinityLive);
ImgLinearRefineRelation(head);
ImgLinearExtractNextStateCase(head);
ImgLinearExtractSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddNextStateCase(head);
ImgLinearRefineRelation(head);
}
includeZeroGain = 1;
varLimit = 200;
affinityLimit = 0.7;
andExistLimit = 5000;
fprintf(stdout, "Heap DAL affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
bOptimize |= ImgLinearClusterUsingHeap(head, affinityLimit, andExistLimit,
varLimit, optDir, 0,
ImgLinearHeapCompareDeadAffinityLive);
ImgLinearRefineRelation(head);
ImgLinearExtractNextStateCase(head);
ImgLinearExtractSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddNextStateCase(head);
ImgLinearRefineRelation(head);
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
includeZeroGain = 1;
affinityLimit = 0.4;
andExistLimit = 5000;
fprintf(stdout, "Heap DAL affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
bOptimize |= ImgLinearClusterUsingHeap(head, affinityLimit, andExistLimit,
varLimit, optDir, 0,
ImgLinearHeapCompareDeadAffinityLive);
ImgLinearRefineRelation(head);
ImgLinearExtractNextStateCase(head);
ImgLinearExtractSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddNextStateCase(head);
ImgLinearRefineRelation(head);
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
affinityLimit = 0.0;
andExistLimit = 5000;
fprintf(stdout, "Heap DLA affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
bOptimize |= ImgLinearClusterUsingHeap(head, affinityLimit, andExistLimit,
varLimit, optDir, 0,
ImgLinearHeapCompareDeadLiveAffinity);
ImgLinearRefineRelation(head);
ImgLinearExtractNextStateCase(head);
ImgLinearRefineRelation(head);
ImgLinearAddNextStateCase(head);
ImgLinearRefineRelation(head);
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
if(head->computeRange) {
includeZeroGain = 0;
affinityLimit = 0.0;
andExistLimit = andExistLimit;
varLimit = MAXINT;
head->bddLimit = head->bddLimit * 2;
fprintf(stdout, "Heap DLA affinity %3f, andExist %d\n",
affinityLimit, andExistLimit);
while(1) {
bOptimize |= ImgLinearClusterUsingHeap(head, affinityLimit, andExistLimit,
varLimit, optDir, 1,
ImgLinearHeapCompareDeadLiveAffinity);
ImgLinearRefineRelation(head);
if(ImgCheckRangeTestAndOverapproximate(head)) break;
}
ImgCountOnsetDisjunctiveArray(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
}
return(bOptimize);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearClusteringByConstraints | ( | Relation_t * | head, |
| int | includeZeroGain, | ||
| int | varLimit, | ||
| int | clusterLimit, | ||
| int | gainLimit, | ||
| double | affinityLimit, | ||
| int | andExistLimit, | ||
| int | bddLimit, | ||
| int | useFailureHistory, | ||
| int(*)(const void *, const void *) | compare_func | ||
| ) |
Function********************************************************************
Synopsis [Apply clustering with given priority function]
Description [Apply clustering with given priority function]
SideEffects []
SeeAlso []
Definition at line 1480 of file imgLinear.c.
{
int size, begin, end;
int **nDead, **nVar, **nFailure;
int *dirtyBit;
int i, j, k, l, index;
int start, preGain;
int nClusterArray;
VarLife_t **varArray, *var;
Cluster_t **clusterArray, *clu;
Conjunct_t *conjunct;
bdd_t *clusteredRelation;
int fail_flag;
int from, to, splitable;
varArray = head->varArray;
size = head->nSize;
nDead = ALLOC(int *, size);
nVar = ALLOC(int *, size);
nFailure = 0;
if(useFailureHistory)
nFailure = ALLOC(int *, size);
for(i=0; i<size; i++) {
nDead[i] = ALLOC(int, size);
nVar[i] = ALLOC(int, size);
if(useFailureHistory)
nFailure[i] = ALLOC(int, size);
memset(nDead[i], 0, sizeof(int)*size);
memset(nVar[i], 0, sizeof(int)*size);
if(useFailureHistory)
memset(nFailure[i], 0, sizeof(int)*size);
}
index = 0;
for(l=0; l<head->nVarArray; l++) {
var = varArray[l];
begin = var->from;
end = var->to;
if(var->stateVar == 1) begin = -1;
else if(var->stateVar == 2) end = size;
for(i=0; i<=begin; i++)
for(j=end; j<size; j++)
nDead[i][j]++;
for(i=0; i<=var->effTo; i++) {
if(i > var->effFrom) {
for(k=0; k<var->nSize; k++) {
index = var->relArr[k];
if(index == MAXINT) continue;
if(index == MAXINT-1) continue;
if(index >= i) break;
}
if(index < i) start = var->effTo;
else start = index;
}
else {
start = var->effFrom;
}
for(j=start; j<size; j++)
nVar[i][j]++;
}
}
for(i=0; i<size; i++) {
end = (size-1) < i+clusterLimit ? size-1 : i+clusterLimit;
if(i==end) continue;
if(nDead[i][end] == 0 && !includeZeroGain) continue;
preGain = -1;
for(j=end; j>=i; j--) {
if(varLimit < nVar[i][j]-nDead[i][j]) continue;
if(gainLimit < nDead[i][j]) continue;
if(preGain != -1) {
if(nDead[i][j] < preGain && nDead[i][j] >= 0) {
to = j+1;
for(from=i; from < to; from++) {
if(nDead[from+1][to] != nDead[from][to]) break;
}
if(from != i) continue;
splitable = 0;
for(k=from; k<to; k++) {
if(nDead[from][to] == (nDead[from][k] + nDead[k+1][to])) {
splitable = 1;
break;
}
}
if(!splitable)
ImgLinearInsertClusterCandidate(head, from, to,
nDead[from][to], nVar[from][to], affinityLimit);
}
}
if(nDead[i][j] == 0) {
if(includeZeroGain)
ImgLinearInsertClusterCandidate(head, i, i+1,
nDead[i][i+1], nVar[i][i+1], affinityLimit);
break;
}
preGain = nDead[i][j];
}
}
if(head->nClusterArray == 0) return;
qsort(head->clusterArray, head->nClusterArray, sizeof(Cluster_t *),
compare_func);
dirtyBit = ALLOC(int, size);
memset(dirtyBit, 0, sizeof(int)*size);
clusterArray = head->clusterArray;
nClusterArray = head->nClusterArray;
for(i=0; i<nClusterArray; i++) {
clu = clusterArray[i];
if(useFailureHistory) {
if(nFailure[clu->from][clu->to] == 1) continue;
}
fail_flag = 0;
for(j=clu->from; j<=clu->to; j++) {
if(dirtyBit[j] == 1) {
fail_flag = 1;
continue;
}
}
if(fail_flag) continue;
clusteredRelation = ImgLinearClusteringSmooth(head, clu,
nFailure, andExistLimit, bddLimit);
conjunct = 0;
if(clusteredRelation) {
for(j=clu->from; j<=clu->to; j++) {
conjunct = head->conjunctArray[j];
if(conjunct->relation) bdd_free(conjunct->relation);
conjunct->relation = 0;
}
conjunct->relation = clusteredRelation;
conjunct->bModified = 1;
head->bModified = 1;
for(j=clu->from; j<=clu->to; j++) dirtyBit[j] = 1;
if(head->verbosity >= 5)
fprintf(stdout, "\tClustering Success : %4d -%4d G(%3d) V(%3d) A(%3d)\n",
clu->from, clu->to, clu->nDead, clu->nVar, clu->nAffinity);
}
fflush(stdout);
}
for(i=0; i<nClusterArray; i++)
free(clusterArray[i]);
free(clusterArray);
head->clusterArray = 0;
head->nClusterArray = 0;
for(i=0; i<head->nSize; i++) {
free(nDead[i]);
free(nVar[i]);
if(useFailureHistory)
free(nFailure[i]);
}
free(nDead);
free(nVar);
if(useFailureHistory)
free(nFailure);
return;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| int ImgLinearClusteringIteratively | ( | Relation_t * | head, |
| double | affinityLimit, | ||
| int | andExistLimit, | ||
| int | varLimit, | ||
| int | includeZeroGain, | ||
| int | useFailureHistory, | ||
| Img_OptimizeType | optDir, | ||
| int(*)(const void *, const void *) | compare_func | ||
| ) |
Function********************************************************************
Synopsis [Apply clustering iteratively]
Description [Apply clustering iteratively]
SideEffects []
SeeAlso []
clusterLimit = head->nSize/5; if(clusterLimit < 1) clusterLimit = 3;
Definition at line 1429 of file imgLinear.c.
{
int bddLimit, gainLimit;
int clusterLimit;
int bOptimize;
if(head->nSize < 2) return(0);
bOptimize = 0;
bddLimit = head->bddLimit;
gainLimit = 50;
while(1) {
if(head->nSize < 50)clusterLimit = head->nSize/2;
else clusterLimit = head->nSize/3+1;
if(clusterLimit > 50) clusterLimit = 50;
ImgLinearClusteringByConstraints(head, includeZeroGain, varLimit, clusterLimit,
gainLimit, affinityLimit, andExistLimit,
bddLimit, useFailureHistory, compare_func);
if(head->bModified) {
ImgLinearRefineRelation(head);
bOptimize |= ImgLinearOptimizeAll(head, optDir, 0);
}
else break;
}
return(bOptimize);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static bdd_t * ImgLinearClusteringPairSmooth | ( | Relation_t * | head, |
| Cluster_t * | clu, | ||
| int ** | failureHistory, | ||
| int | andExistLimit, | ||
| int | bddLimit | ||
| ) | [static] |
Function********************************************************************
Synopsis [Clustering pair of transition relation]
Description [Clustering pair of transition relation]
SideEffects []
SeeAlso []
Definition at line 1998 of file imgLinear.c.
{
int *varType, *supList;
array_t *smoothArray;
Conjunct_t *conjunct;
bdd_t *totalRelation;
bdd_t *fromRelation, *toRelation;
bdd_t *varBdd;
int i, j, k, tempSize;
int id, tid, effTo;
st_table *deadTable;
VarLife_t *var;
varType = head->varType;
smoothArray = array_alloc(bdd_t *, 0);
for(effTo = clu->to+1; effTo<head->nSize; effTo++) {
conjunct = head->conjunctArray[effTo];
if(conjunct && conjunct->relation) break;
}
effTo--;
deadTable = st_init_table(st_numcmp, st_numhash);
for(i=clu->from; i<=clu->to; i++) {
conjunct = head->conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
supList = conjunct->supList;
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
if(varType[id] == 1 || varType[id] == 2) continue;
if(st_lookup(deadTable, (char *)(long)id, &tid)) continue;
var = head->varArray[head->varArrayMap[id]];
if(clu->from <= var->from && var->to <= effTo) {
st_insert(deadTable, (char *)(long)id, (char *)(long)id);
varBdd = bdd_get_variable(head->mgr, var->id);
array_insert_last(bdd_t *, smoothArray, varBdd);
}
}
}
st_free_table(deadTable);
conjunct = head->conjunctArray[clu->from];
fromRelation = conjunct->relation;
toRelation = 0;
for(i=clu->from+1; i<=clu->to; i++) {
conjunct = head->conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
toRelation = conjunct->relation;
break;
}
if(toRelation == 0) {
mdd_array_free(smoothArray);
return(0);
}
if(fromRelation == 0 && toRelation == 0) return(0);
if(fromRelation == 0) {
totalRelation = bdd_dup(toRelation);
}
else if(toRelation == 0) {
totalRelation = bdd_dup(fromRelation);
}
else {
totalRelation = bdd_and_smooth_with_limit(fromRelation, toRelation,
smoothArray, andExistLimit);
}
if(totalRelation) tempSize = bdd_size(totalRelation);
else tempSize = MAXINT;
if(tempSize > bddLimit) {
if(totalRelation) bdd_free(totalRelation);
totalRelation = 0;
if(failureHistory) {
for(j=0; j<=clu->from; j++)
for(k=i; k<head->nSize; k++)
failureHistory[j][k] = 1;
}
if(head->verbosity >= 3)
fprintf(stdout, "Clustering Failure : %4d-%4d G(%3d) V(%3d) A(%3d) At %3d S(%d)\n",
clu->from, clu->to, clu->nDead, clu->nVar, clu->nAffinity, i, tempSize);
}
mdd_array_free(smoothArray);
return(totalRelation);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static bdd_t * ImgLinearClusteringSmooth | ( | Relation_t * | head, |
| Cluster_t * | clu, | ||
| int ** | failureHistory, | ||
| int | andExistLimit, | ||
| int | bddLimit | ||
| ) | [static] |
Function********************************************************************
Synopsis [Apply clustering while quantifying the variables that are isolated]
Description [Apply clustering while quantifying the variables that are isolated]
SideEffects []
SeeAlso []
Definition at line 1768 of file imgLinear.c.
{
int *varType, *supList;
array_t *smoothVarBddArray, *smoothArray;
Conjunct_t *conjunct;
bdd_t *relation, *totalRelation;
bdd_t *varBdd, *tempRelation;
int i, j, k, tempSize, failFlag;
int id, tid;
st_table *deadTable;
VarLife_t *var;
varType = head->varType;
smoothVarBddArray = array_alloc(array_t *, 0);
for(i=clu->from; i<=clu->to; i++) {
smoothArray = array_alloc(bdd_t *, 0);
array_insert_last(array_t *, smoothVarBddArray, smoothArray);
}
deadTable = st_init_table(st_numcmp, st_numhash);
for(i=clu->from; i<=clu->to; i++) {
conjunct = head->conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
supList = conjunct->supList;
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
if(varType[id] == 1 || varType[id] == 2) continue;
if(st_lookup(deadTable, (char *)(long)id, &tid)) continue;
var = head->varArray[head->varArrayMap[id]];
if(clu->from <= var->from && var->to <= clu->to) {
st_insert(deadTable, (char *)(long)id, (char *)(long)id);
smoothArray = array_fetch(array_t *, smoothVarBddArray,
var->to-clu->from);
varBdd = bdd_get_variable(head->mgr, var->id);
array_insert_last(bdd_t *, smoothArray, varBdd);
}
}
}
st_free_table(deadTable);
totalRelation = bdd_one(head->mgr);
failFlag = 0;
for(i=clu->from; i<=clu->to; i++) {
conjunct = head->conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
relation = conjunct->relation;
smoothArray = array_fetch(array_t *, smoothVarBddArray, i-clu->from);
tempRelation = bdd_and_smooth_with_limit(totalRelation, relation,
smoothArray, andExistLimit);
if(tempRelation) tempSize = bdd_size(tempRelation);
else tempSize = MAXINT;
if(tempSize > bddLimit) {
bdd_free(totalRelation);
if(tempRelation) bdd_free(tempRelation);
totalRelation = 0;
if(failureHistory) {
for(j=0; j<=clu->from; j++)
for(k=i; k<head->nSize; k++)
failureHistory[j][k] = 1;
}
if(head->verbosity >= 3)
fprintf(stdout, "Clustering Failure : %4d-%4d G(%3d) V(%3d) A(%3d) At %3d S(%d)\n",
clu->from, clu->to, clu->nDead, clu->nVar, clu->nAffinity, i, tempSize);
break;
}
bdd_free(totalRelation);
totalRelation = tempRelation;
}
ImgLinearFreeSmoothArray(smoothVarBddArray);
if(failFlag) return(0);
else return(totalRelation);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearClusterRelationArray | ( | mdd_manager * | mgr, |
| ImgFunctionData_t * | functionData, | ||
| array_t * | relationArray, | ||
| Img_DirectionType | direction, | ||
| array_t ** | clusteredRelationArrayPtr, | ||
| array_t ** | arraySmoothVarBddArrayPtr, | ||
| array_t ** | optClusteredRelationArrayPtr, | ||
| array_t ** | optArraySmoothVarBddArrayPtr, | ||
| ImgTrmOption_t * | option | ||
| ) |
Function********************************************************************
Synopsis [Cluster fine grain transition relation]
Description [First the linear arrangement of transition relation that minimze max cut is generated and the iterative clustering is applied on it.]
SideEffects []
SeeAlso []
Definition at line 176 of file imgLinear.c.
{
Img_OptimizeType optDir;
int bGroupStateVariable;
int bOrderVariable;
int includeCS;
int includeNS;
int quantifyCS;
array_t *initRelationArray;
int bOptimize;
Relation_t *head;
bdd_t **smoothCubeArr, **optSmoothCubeArr;
array_t *optRelationArr, *optSmoothVarBddArr;
array_t *relationArr, *smoothVarBddArr;
if(option->linearComputeRange) {
option->linearIncludeCS = 0;
option->linearIncludeNS = 0;
option->linearQuantifyCS = 1;
}
optRelationArr = relationArr = 0;
optSmoothVarBddArr = smoothVarBddArr = 0;
includeCS = option->linearIncludeCS;
includeNS = option->linearIncludeNS;
quantifyCS = option->linearQuantifyCS;
if(option->linearFineGrainFlag) includeNS = 1;
optDir = option->linearOptimize;
bOrderVariable = option->linearOrderVariable;
bGroupStateVariable = option->linearGroupStateVariable;
head = ImgLinearRelationInit(mgr, relationArray,
functionData->domainBddVars, functionData->rangeBddVars,
functionData->quantifyBddVars, option);
if(head->verbosity >= 5) ImgLinearPrintDebugInfo(head);
ImgLinearOptimizeAll(head, Opt_None, 0);
ImgLinearRefineRelation(head);
initRelationArray = ImgLinearExtractRelationArrayT(head);
bOptimize = 0;
ImgLinearPrintMatrix(head);
if(bOrderVariable)
ImgLinearVariableOrder(head, "VarOrder", !bGroupStateVariable);
if((direction==0) || (direction==2)) {
ImgLinearPrintMatrix(head);
bOptimize |= ImgLinearOptimizeAll(head, optDir, 0);
ImgLinearRefineRelation(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearConjunctOrderMainCC(head, 0);
ImgLinearPrintMatrix(head);
bOptimize |= ImgLinearClustering(head, optDir);
ImgLinearRefineRelation(head);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintTransitionInfo(head);
ImgLinearPrintMatrix(head);
ImgLinearPrintMatrixFull(head, 2);
if(bOptimize) {
optRelationArr = ImgLinearExtractRelationArrayT(head);
optSmoothCubeArr = ALLOC(bdd_t *, head->nSize+1);
optSmoothVarBddArr = ImgLinearMakeSmoothVarBdd(head, optSmoothCubeArr);
}
else {
optRelationArr = ImgLinearExtractRelationArrayT(head);
optSmoothCubeArr = ALLOC(bdd_t *, head->nSize+1);
optSmoothVarBddArr = ImgLinearMakeSmoothVarBdd(head, optSmoothCubeArr);
relationArr = 0;
smoothVarBddArr = 0;
}
ImgLinearPrintDebugInfo(head);
ImgLinearRelationQuit(head);
if(bOptimize) {
fprintf(vis_stdout, "Get Schedules without optimization\n");
head = ImgLinearRelationInit(mgr, initRelationArray,
functionData->domainBddVars, functionData->rangeBddVars,
functionData->quantifyBddVars, option);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearConjunctOrderMainCC(head, 0);
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
ImgLinearClustering(head, Opt_None);
relationArr = ImgLinearExtractRelationArrayT(head);
smoothCubeArr = ALLOC(bdd_t *, head->nSize+1);
smoothVarBddArr = ImgLinearMakeSmoothVarBdd(head, smoothCubeArr);
ImgLinearSetEffectiveNumberOfStateVariable(head, 0, 0, 0);
ImgLinearPrintDebugInfo(head);
ImgLinearPrintTransitionInfo(head);
ImgLinearRelationQuit(head);
}
if(optClusteredRelationArrayPtr)
*optClusteredRelationArrayPtr = optRelationArr;
else
mdd_array_free(optRelationArr);
if(optArraySmoothVarBddArrayPtr)
*optArraySmoothVarBddArrayPtr = optSmoothVarBddArr;
else if(optSmoothVarBddArr)
ImgLinearFreeSmoothArray(optSmoothVarBddArr);
if(clusteredRelationArrayPtr)
*clusteredRelationArrayPtr = relationArr;
else
mdd_array_free(relationArr);
if(arraySmoothVarBddArrayPtr)
*arraySmoothVarBddArrayPtr = smoothVarBddArr;
else if(smoothVarBddArr)
ImgLinearFreeSmoothArray(smoothVarBddArr);
}
if((direction==1) || (direction==2)) {
}
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearClusterSameSupportSet | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Cluster the relation that has same support set]
Description [Cluster the relation that has same support set]
SideEffects []
SeeAlso []
Definition at line 4421 of file imgLinear.c.
{
int i, j, index, id;
int *supList;
Conjunct_t *conjunct, *base;
int *varType;
Conjunct_t **conjunctArray, **newConjunctArray;
head->nTotalCluster = 0;
varType = head->varType;
conjunctArray = head->conjunctArray;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
supList = conjunct->supList;
conjunct->dummy = 0;
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
if(varType[id] == 2) continue;
conjunct->dummy += id;
}
conjunct->dummy *= (conjunct->nSize-conjunct->nRange);
conjunct->dummy -= conjunct->nDomain;
}
qsort(conjunctArray, head->nSize, sizeof(Conjunct_t *),
ImgLinearCompareConjunctDummy);
for(i=0; i<head->nSize; i++) {
base = head->conjunctArray[i];
for(j=i+1; j<head->nSize; j++) {
conjunct = head->conjunctArray[j];
if(base->dummy != conjunct->dummy) {
if(j == i+1) break;
ImgLinearFindSameSupportConjuncts(head, i, j-1);
i = j-1;
break;
}
}
}
if(head->bModified) {
conjunctArray = head->conjunctArray;
newConjunctArray = ALLOC(Conjunct_t *, head->nSize);
for(index=0, i=0; i<head->nSize; i++) {
conjunct = conjunctArray[i];
if(conjunct == 0) continue;
newConjunctArray[index++] = conjunct;
}
free(conjunctArray);
head->conjunctArray = newConjunctArray;
head->nSize = index;
head->bModified = 1;
head->bRefineVarArray = 1;
}
conjunctArray = head->conjunctArray;
qsort(conjunctArray, head->nSize, sizeof(Conjunct_t *), ImgLinearCompareConjunctIndex);
if(head->bModified) {
conjunctArray = head->conjunctArray;
for(i=0; i<head->nSize; i++) {
conjunct = conjunctArray[i];
if(conjunct == 0) continue;
conjunct->index = i;
}
}
fprintf(stdout, "NOTICE : %d conjunct clustered\n", head->nTotalCluster);
head->nTotalCluster = 0;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearClusterUsingHeap | ( | Relation_t * | head, |
| double | affinityLimit, | ||
| int | andExistLimit, | ||
| int | varLimit, | ||
| Img_OptimizeType | optDir, | ||
| int | rangeFlag, | ||
| int(*)(const void *, const void *) | compare_func | ||
| ) | [static] |
Function********************************************************************
Synopsis [ Apply clustering with priority queue]
Description [ Apply clustering with priority queue]
SideEffects []
SeeAlso []
need to consider statevariable optimization
for(i=0; i<heap->nitems; i++) { tclu = (Cluster_t *)(heap->slots[i].item); if(tclu->from <= clu->from && clu->from <= tclu->to) tclu->flag = 1; if(tclu->from <= clu->to && clu->to <= tclu->to) tclu->flag = 1; }
Definition at line 1871 of file imgLinear.c.
{
Cluster_t *clu;
int j;
Conjunct_t *conjunct;
bdd_t *clusteredRelation;
Heap_t *heap;
int bOptimize;
long dummy;
bOptimize = 0;
ImgLinearBuildInitialCandidate(head, affinityLimit, varLimit, rangeFlag, compare_func);
heap = head->clusterHeap;
while(Heap_HeapExtractMin(heap, &clu, &dummy)) {
if(clu->flag) {
free(clu);
continue;
}
if(clu->to >= head->nSize) {
free(clu);
continue;
}
clusteredRelation = ImgLinearClusteringPairSmooth(head, clu,
0, andExistLimit, head->bddLimit);
if(clusteredRelation) {
if(head->verbosity >= 5)
fprintf(stdout, "\tClustering Success : %4d -%4d G(%3d) V(%3d) A(%3d)\n",
clu->from, clu->to, clu->nDead, clu->nVar, clu->nAffinity);
fflush(stdout);
for(j=clu->from+1; j<=clu->to; j++) {
conjunct = head->conjunctArray[j];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
bdd_free(conjunct->relation);
conjunct->relation = 0;
}
conjunct = head->conjunctArray[clu->from];
if(conjunct->relation)
bdd_free(conjunct->relation);
conjunct->relation = clusteredRelation;
conjunct->bModified = 1;
ImgLinearConjunctRefine(head, conjunct);
head->bModified = 1;
head->bRefineVarArray = 1;
Heap_HeapApplyForEachElement(heap, linearCheckRange);
ImgLinearInsertPairClusterCandidate(head, clu->from, affinityLimit, varLimit, rangeFlag);
ImgLinearInsertPairClusterCandidate(head, clu->from-1, affinityLimit, varLimit, rangeFlag);
}
if(clu->nDead && clusteredRelation) {
ImgLinearVariableArrayInit(head);
bOptimize |= ImgLinearOptimizeAll(head, optDir, 0);
}
free(clu);
}
Heap_HeapFree(heap);
head->clusterHeap = 0;
if(head->bModified) {
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
bOptimize |= ImgLinearOptimizeAll(head, optDir, 0);
ImgLinearRefineRelation(head);
if(head->verbosity >= 5)
ImgLinearPrintMatrix(head);
}
return(bOptimize);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareAffinityDeadLive | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3777 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = *(Cluster_t **)c1;
clu2 = *(Cluster_t **)c2;
if(clu1->nAffinity == clu2->nAffinity) {
if(clu1->nDead == clu2->nDead) {
return(clu1->nVar-clu1->nDead > clu2->nVar-clu2->nDead);
}
return(clu1->nDead < clu2->nDead);
}
return(clu1->nAffinity < clu2->nAffinity);
}
| static int ImgLinearCompareConjunctDummy | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3634 of file imgLinear.c.
{
Conjunct_t *con1, *con2;
con1 = *(Conjunct_t **)c1;
con2 = *(Conjunct_t **)c2;
return(con1->dummy > con2->dummy);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareConjunctIndex | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3655 of file imgLinear.c.
{
Conjunct_t *con1, *con2;
con1 = *(Conjunct_t **)c1;
con2 = *(Conjunct_t **)c2;
return(con1->index > con2->index);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareConjunctRangeMinusDomain | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3613 of file imgLinear.c.
{
Conjunct_t *con1, *con2;
con1 = *(Conjunct_t **)c1;
con2 = *(Conjunct_t **)c2;
return(con1->nRange-con1->nDomain > con2->nRange-con2->nDomain);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareConjunctSize | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3677 of file imgLinear.c.
{
Conjunct_t *con1, *con2;
con1 = *(Conjunct_t **)c1;
con2 = *(Conjunct_t **)c2;
if(con1->nSize == con2->nSize)
return(con1->dummy < con2->dummy);
return(con1->nSize < con2->nSize);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareDeadAffinityLive | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3723 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = *(Cluster_t **)c1;
clu2 = *(Cluster_t **)c2;
if(clu1->nDead == clu2->nDead) {
if(clu1->nAffinity == clu2->nAffinity) {
return(clu1->nVar-clu1->nDead > clu2->nVar-clu2->nDead);
}
return(clu1->nAffinity < clu2->nAffinity);
}
return(clu1->nDead < clu2->nDead);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareDeadLiveAffinity | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3750 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = *(Cluster_t **)c1;
clu2 = *(Cluster_t **)c2;
if(clu1->nDead == clu2->nDead) {
if(clu1->nVar-clu1->nDead == clu2->nVar-clu2->nDead) {
return(clu1->nAffinity < clu2->nAffinity);
}
return(clu1->nVar-clu1->nDead > clu2->nVar-clu2->nDead);
}
return(clu1->nDead < clu2->nDead);
}
| static int ImgLinearCompareLiveAffinityDead | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3804 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = *(Cluster_t **)c1;
clu2 = *(Cluster_t **)c2;
if(clu1->nVar-clu1->nDead == clu2->nVar-clu2->nDead) {
if(clu1->nAffinity == clu2->nAffinity) {
return(clu1->nDead < clu2->nDead);
}
return(clu1->nAffinity < clu2->nAffinity);
}
return(clu1->nVar-clu1->nDead > clu2->nVar-clu2->nDead);
}
| static int ImgLinearCompareVarDummyLarge | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3529 of file imgLinear.c.
{
VarLife_t *con1, *con2;
con1 = *(VarLife_t **)c1;
con2 = *(VarLife_t **)c2;
return(con1->dummy < con2->dummy);
}
| static int ImgLinearCompareVarEffFromLarge | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3550 of file imgLinear.c.
{
VarLife_t *con1, *con2;
con1 = *(VarLife_t **)c1;
con2 = *(VarLife_t **)c2;
return(con1->effFrom < con2->effFrom);
}
| static int ImgLinearCompareVarEffFromSmall | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3508 of file imgLinear.c.
{
VarLife_t *con1, *con2;
con1 = *(VarLife_t **)c1;
con2 = *(VarLife_t **)c2;
return(con1->effFrom > con2->effFrom);
}
| static int ImgLinearCompareVarId | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3571 of file imgLinear.c.
{
VarLife_t *con1, *con2;
con1 = *(VarLife_t **)c1;
con2 = *(VarLife_t **)c2;
return(con1->id < con2->id);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareVarIndex | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3701 of file imgLinear.c.
{
VarLife_t *v1, *v2;
v1 = *(VarLife_t **)c1;
v2 = *(VarLife_t **)c2;
return(v1->index < v2->index);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearCompareVarSize | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3592 of file imgLinear.c.
{
VarLife_t *con1, *con2;
con1 = *(VarLife_t **)c1;
con2 = *(VarLife_t **)c2;
return(con1->nSize > con2->nSize);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearComputeLifeTime | ( | Relation_t * | head, |
| double * | paal, | ||
| double * | patl | ||
| ) | [static] |
Function********************************************************************
Synopsis [Compute life time of variables]
Description [Compute life time of variables]
SideEffects []
SeeAlso []
Definition at line 3381 of file imgLinear.c.
{
double aal, atl;
int i;
VarLife_t **varArray, *var;
aal = atl = 0.0;
varArray = head->varArray;
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
aal += var->effTo - var->effFrom;
if(var->stateVar == 1) atl += var->to+1;
else if(var->stateVar == 2) atl += head->nSize+1 - var->from;
else atl += var->to - var->from;
}
atl = atl / (double)(head->nVarArray * head->nSize);
aal = aal / (double)(head->nVarArray * head->nSize);
*paal = aal;
*patl = atl;
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearConjunctArrayRefine | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Refine conjunction array to fill the empty slot.]
Description [Refine conjunction array to fill the empty slot.]
SideEffects []
SeeAlso []
Definition at line 4202 of file imgLinear.c.
{
int nSize, i;
int index;
Conjunct_t **conjunctArray, **newConjunctArray;
Conjunct_t *conjunct;
if(head->bModified == 0) return;
conjunctArray = head->conjunctArray;
newConjunctArray = ALLOC(Conjunct_t *, head->nSize+1);
nSize = head->nSize;
for(index=0, i=0; i<nSize; i++) {
conjunct = conjunctArray[i];
if(conjunct == 0) {
head->bRefineVarArray = 1;
continue;
}
if(conjunct->relation == 0) {
ImgLinearConjunctQuit(conjunct);
head->bRefineVarArray = 1;
continue;
}
if(conjunct->bModified) {
ImgLinearConjunctRefine(head, conjunct);
head->bRefineVarArray = 1;
}
if(conjunct == 0) continue;
if(conjunct->relation == 0) {
ImgLinearConjunctQuit(conjunct);
continue;
}
newConjunctArray[index] = conjunct;
conjunct->index = index++;
}
newConjunctArray[index] = 0;
free(conjunctArray);
head->conjunctArray = newConjunctArray;
head->nSize = index;
head->bModified = 0;
return;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearConjunctionOrder | ( | Relation_t * | head, |
| char * | baseName, | ||
| int | refineFlag | ||
| ) | [static] |
Function********************************************************************
Synopsis [Generate linear arrangement with CAPO]
Description [Generate linear arrangement with CAPO]
SideEffects []
SeeAlso []
Definition at line 2400 of file imgLinear.c.
{
Conjunct_t *conjunct;
int i, j;
if(head->nSize < 2) return;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
conjunct->dummy = 0;
for(j=0; j<conjunct->nSize; j++)
conjunct->dummy += conjunct->supList[j];
}
qsort(head->conjunctArray, head->nSize, sizeof(Conjunct_t *),
ImgLinearCompareConjunctSize);
head->bRefineVarArray = 1;
ImgLinearRefineRelation(head);
ImgLinearCAPOInterfaceConjunctNodes(head, baseName);
ImgLinearCAPOInterfaceConjunctNet(head, baseName);
ImgLinearCAPOInterfaceConjunctScl(head, baseName);
ImgLinearCAPOInterfaceConjunctPl(head, baseName);
ImgLinearCAPOInterfaceAux(head, baseName);
ImgLinearCAPORun("MetaPlacer", baseName, 0);
ImgLinearCAPOReadConjunctOrder(head, baseName);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearConjunctOrderMain | ( | Relation_t * | head, |
| int | bRefineConjunctOrder | ||
| ) | [static] |
Function********************************************************************
Synopsis [ Main function of linear arrangement]
Description [ Main function of linear arrangement]
SideEffects []
SeeAlso []
head->includeCS = 0; ImgLinearConjunctionOrder(head, "FirstCon", bRefineConjunctOrder); ImgLinearComputeLifeTime(head, &aal, &atl); ImgLinearPrintMatrixFull(head, 0); head->includeCS = 1; tempConjunctArray = ALLOC(Conjunct_t *, head->nSize+1); for(i=0; i<head->nSize; i++) tempConjunctArray[i] = head->conjunctArray[i]; tempAal = aal; tempAtl = atl; fprintf(stdout, "NOTICE : aal %.3f atl %.3f\n", aal, atl); ImgLinearPrintMatrix(head);
if(tempAal < aal) { if( (aal-tempAal) > (tempAtl-atl)*2.0 ) { free(head->conjunctArray); head->conjunctArray = tempConjunctArray; head->bModified = 1; head->bRefineVarArray = 1; ImgLinearRefineRelation(head); fprintf(stdout, "includeCS = 0 is taken\n"); } else { fprintf(stdout, "includeCS = 1 is taken\n"); } } else { fprintf(stdout, "includeCS = 1 is taken\n"); }
Definition at line 2342 of file imgLinear.c.
{
double aal, atl;
ImgLinearConjunctionOrder(head, "SecondCon", bRefineConjunctOrder);
ImgLinearComputeLifeTime(head, &aal, &atl);
ImgLinearPrintMatrixFull(head, 1);
fprintf(stdout, "NOTICE : aal %.3f atl %.3f\n", aal, atl);
ImgLinearPrintMatrix(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearConjunctOrderMainCC | ( | Relation_t * | head, |
| int | refineFlag | ||
| ) |
AutomaticEnd
Function********************************************************************
Synopsis [Order the fine grain transition relation.]
Description [First find the connected component and apply ordering procedure for each component.]
SideEffects []
SeeAlso []
Definition at line 907 of file imgLinear.c.
{
Conjunct_t **connectedArray;
int i, index;
char baseName[1024];
ImgLinearPrintMatrix(head);
ImgLinearExtractNextStateCase(head);
ImgLinearClusterSameSupportSet(head);
ImgLinearRefineRelation(head);
ImgLinearExtractSingletonCase(head);
ImgLinearRefineRelation(head);
ImgLinearConnectedComponent(head);
for(i=0; i<head->nConnectedComponent; i++) {
connectedArray = head->connectedComponent[i];
for(index=0; (long)(connectedArray[index])>0; index++);
head->conjunctArray = connectedArray;
head->nSize = index;
head->bModified = 1;
head->bRefineVarArray = 1;
qsort(head->conjunctArray, head->nSize, sizeof(Conjunct_t *),
ImgLinearCompareConjunctSize);
ImgLinearVariableArrayInit(head);
ImgLinearConjunctOrderMain(head, 0);
fprintf(stdout, "NOTICE : %d'th connected component\n", i);
head->connectedComponent[i] = head->conjunctArray;
head->conjunctArray = 0;
}
ImgLinearBuildConjunctArrayWithQuotientCC(head);
ImgLinearAddSingletonCase(head);
ImgLinearAddNextStateCase(head);
ImgLinearExpandSameSupportSet(head);
ImgLinearRefineRelation(head);
ImgLinearRefineRelation(head);
ImgLinearPrintMatrix(head);
sprintf(baseName, "profile");
ImgLinearPrintVariableProfile(head, baseName);
ImgLinearPrintMatrixFull(head, 2);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearConjunctQuit | ( | Conjunct_t * | conjunct | ) | [static] |
Function********************************************************************
Synopsis [Free conjunct_t data structure]
Description [Free conjunct_t data structure]
SideEffects []
SeeAlso []
Definition at line 4605 of file imgLinear.c.
{
if(conjunct->supList) {
free(conjunct->supList);
conjunct->supList = 0;
}
if(conjunct->relation) {
bdd_free(conjunct->relation);
conjunct->relation = 0;
}
free(conjunct);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearConjunctRefine | ( | Relation_t * | head, |
| Conjunct_t * | conjunct | ||
| ) | [static] |
Function********************************************************************
Synopsis [Refine conjunction array to fill the empty slot.]
Description [Refine conjunction array to fill the empty slot.]
SideEffects []
SeeAlso []
Definition at line 4143 of file imgLinear.c.
{
int i, id, listSize, *supList;
int *varType;
if(conjunct->relation == 0) {
if(conjunct->supList) free(conjunct->supList);
conjunct->supList = 0;
return;
}
if(!conjunct->bModified) return;
if(conjunct->supList) {
free(conjunct->supList);
conjunct->supList = 0;
}
supList = ImgLinearGetSupportBddId(head->mgr, conjunct->relation, &listSize);
if(listSize == 0) {
conjunct->relation = 0;
if(conjunct->supList) {
free(conjunct->supList);
conjunct->supList = 0;
}
return;
}
else {
varType = head->varType;
conjunct->supList = supList;
conjunct->nSize = listSize;
conjunct->bModified = 0;
conjunct->nDomain = 0;
conjunct->nRange = 0;
conjunct->nQuantify = 0;
conjunct->from = 0;
conjunct->to = 0;
conjunct->dummy = 0;
for(i=0; i<listSize; i++) {
id = supList[i];
if(varType[id] == 1) conjunct->nDomain++;
else if(varType[id] == 2) conjunct->nRange++;
else if(varType[id] == 3) conjunct->nQuantify++;
}
head->bModified = 1;
}
return;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearConnectedComponent | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Find the connected component from fine grain transition relation]
Description [Find the connected component from fine grain transition relation]
SideEffects []
SeeAlso []
singleton case
connected component case
singleton case
connected component case
Definition at line 1093 of file imgLinear.c.
{
Conjunct_t *conjunct;
Conjunct_t **connectedArray;
int i, j, index, cc_index;
int pre_index, pre_cc_index;
ImgLinearVariableArrayInit(head);
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
conjunct->dummy = 0;
}
cc_index = 0;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
if(conjunct->dummy) continue;
cc_index++;
ImgLinearFindConnectedComponent(head, conjunct, cc_index);
}
qsort(head->conjunctArray, head->nSize, sizeof(Conjunct_t *),
ImgLinearCompareConjunctDummy);
head->nConnectedComponent = 0;
pre_index = 0;
pre_cc_index = 1;
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
if(conjunct->dummy != pre_cc_index) {
if((i-pre_index) == 1) {
head->singletonArray = ImgLinearAddConjunctIntoArray(
head->singletonArray, &(head->nSingletonArray),
head->conjunctArray[i-1]);
}
else {
if(head->nConnectedComponent)
head->connectedComponent = REALLOC(Conjunct_t **, head->connectedComponent, head->nConnectedComponent+1);
else
head->connectedComponent = ALLOC(Conjunct_t **, head->nConnectedComponent+1);
connectedArray = ALLOC(Conjunct_t *, (i-pre_index+2));
head->connectedComponent[head->nConnectedComponent++] = connectedArray;
for(index=0, j=pre_index; j<i; j++)
connectedArray[index++] = head->conjunctArray[j];
connectedArray[index] = 0;
}
pre_cc_index = conjunct->dummy;
pre_index = i;
}
}
if((i-pre_index) == 1) {
head->singletonArray = ImgLinearAddConjunctIntoArray(
head->singletonArray, &(head->nSingletonArray),
head->conjunctArray[i-1]);
}
else {
if(head->nConnectedComponent)
head->connectedComponent = REALLOC(Conjunct_t **,
head->connectedComponent, head->nConnectedComponent+1);
else
head->connectedComponent = ALLOC(Conjunct_t **,
head->nConnectedComponent+1);
connectedArray = ALLOC(Conjunct_t *, (i-pre_index+2));
head->connectedComponent[head->nConnectedComponent++] = connectedArray;
for(index=0, j=pre_index; j<i; j++)
connectedArray[index++] = head->conjunctArray[j];
connectedArray[index] = 0;
}
free(head->conjunctArray);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearExpandSameSupportSet | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Find transtion relations that have smae support set]
Description [Find transtion relations that have smae support set and use this information when finding linear arragement]
SideEffects []
SeeAlso []
Definition at line 4376 of file imgLinear.c.
{
int index, i, j;
Conjunct_t **newConjunctArray;
Conjunct_t *conjunct;
newConjunctArray = ALLOC(Conjunct_t *, head->nSize);
for(index=0, i=0; i<head->nSize; i++) {
if(index >= head->nSize)
newConjunctArray = REALLOC(Conjunct_t *, newConjunctArray, index+1);
conjunct = head->conjunctArray[i];
newConjunctArray[index++] = conjunct;
if(conjunct->nCluster) {
for(j=0; j<conjunct->nCluster; j++) {
if(index >= head->nSize)
newConjunctArray = REALLOC(Conjunct_t *, newConjunctArray, index+1);
newConjunctArray[index++] = conjunct->clusterArray[j];
}
head->bModified = 1;
head->bRefineVarArray = 1;
conjunct->nCluster = 0;
if(conjunct->clusterArray) {
free(conjunct->clusterArray);
conjunct->clusterArray = 0;
}
}
}
free(head->conjunctArray);
head->conjunctArray = newConjunctArray;
head->nSize = index;
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearExtractNextStateCase | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Extract the relation that contains next state varaibles only]
Description [Extract the relation that contains next state varaibles only]
SideEffects []
SeeAlso []
Definition at line 1351 of file imgLinear.c.
{
int i, index, flag;
Conjunct_t *conjunct;
Conjunct_t **nextStateArray;
index = 0;
flag = 1;
nextStateArray = ALLOC(Conjunct_t *, index+1);
for(index=0, i=head->nSize-1; i>=0; i--) {
conjunct = head->conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->nSize == conjunct->nRange && flag == 0) {
nextStateArray[index++] = conjunct;
nextStateArray = REALLOC(Conjunct_t *, nextStateArray, index+1);
head->conjunctArray[i] = 0;
head->bModified = 1;
}
if(conjunct->nSize != conjunct->nRange) flag = 0;
}
head->nextStateArray = nextStateArray;
head->nNextStateArray = index;
ImgLinearRefineRelation(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| bdd_t** ImgLinearExtractRelationArray | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Create relation array for image computation from Relation_t data structure]
Description [Create relation array for image computation from Relation_t data structure]
SideEffects []
SeeAlso []
Definition at line 526 of file imgLinear.c.
{
int i;
bdd_t **relationArray;
Conjunct_t *conjunct;
relationArray = ALLOC(bdd_t *, head->nSize+1);
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
relationArray[i] = bdd_dup(conjunct->relation);
}
relationArray[i] = 0;
return(relationArray);
}
| array_t* ImgLinearExtractRelationArrayT | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Create relation array for image computation from Relation_t data structure]
Description [Create relation array for image computation from Relation_t data structure]
SideEffects []
SeeAlso []
Definition at line 552 of file imgLinear.c.
{
int i;
array_t *relationArray;
Conjunct_t *conjunct;
relationArray = array_alloc(bdd_t *, head->nSize);
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
array_insert_last(bdd_t *, relationArray, bdd_dup(conjunct->relation));
}
return(relationArray);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearExtractSingletonCase | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Extract the relation contains only one next state varaible]
Description [Extract the relation contains only one next state varaible]
SideEffects []
SeeAlso []
Definition at line 1225 of file imgLinear.c.
{
int i, j, k, id, index;
VarLife_t **varArray, *var;
int nSingleton;
int *supList, *varType;
Conjunct_t *conjunct, **newSingletonArray;
int *singletonArray;
varType = head->varType;
varArray = head->varArray;
nSingleton = 0;
singletonArray = ALLOC(int , nSingleton+1);
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(var->stateVar == 2) continue;
if(var->stateVar == 1) {
if(var->nSize - head->includeCS > 1) continue;
}
else {
if(var->nSize > 1) continue;
}
for(j=0; j<var->nSize; j++) {
if(var->relArr[j] == MAXINT) continue;
if(var->relArr[j] == MAXINT-1) continue;
singletonArray[nSingleton++] = var->relArr[j];
singletonArray = REALLOC(int , singletonArray, nSingleton+1);
}
}
for(i=0; i<nSingleton; i++) {
conjunct = head->conjunctArray[singletonArray[i]];
conjunct->bSingleton = 1;
}
for(i=0; i<nSingleton; i++) {
conjunct = head->conjunctArray[singletonArray[i]];
if(conjunct->nRange == conjunct->nSize-conjunct->nRange &&
conjunct->nSize-conjunct->nRange != 1) {
conjunct->bSingleton = 0;
continue;
}
supList = conjunct->supList;
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
if(varType[id] == 2) continue;
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] : 0;
if(!var) continue;
if(var->stateVar == 1) {
if(var->nSize - head->includeCS == 1) continue;
}
else {
if(var->nSize == 1) continue;
}
for(k=0; k<var->nSize; k++) {
index = var->relArr[k];
if(index == MAXINT) continue;
if(index == MAXINT-1) continue;
(head->conjunctArray[index])->bSingleton = 0;
}
}
}
newSingletonArray = ALLOC(Conjunct_t *, nSingleton);
for(index=0, i=0; i<nSingleton; i++) {
conjunct = head->conjunctArray[singletonArray[i]];
if(conjunct == 0) continue;
if(conjunct->bSingleton) {
newSingletonArray[index++] = conjunct;
head->conjunctArray[singletonArray[i]] = 0;
}
}
head->nSingletonArray = index;
free(singletonArray);
qsort(newSingletonArray, head->nSingletonArray,
sizeof(Conjunct_t *), ImgLinearCompareConjunctRangeMinusDomain);
head->singletonArray = newSingletonArray;
if(head->nSingletonArray) {
head->bModified = 1;
head->bRefineVarArray = 1;
}
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearFindConnectedComponent | ( | Relation_t * | head, |
| Conjunct_t * | conjunct, | ||
| int | cc_index | ||
| ) |
Function********************************************************************
Synopsis [Find the connected component from fine grain transition relation]
Description [Find the connected component from fine grain transition relation]
SideEffects []
SeeAlso []
Definition at line 1181 of file imgLinear.c.
{
VarLife_t *var;
int *supList;
int id, size;
int index, i, j;
int *relArr;
Conjunct_t *tConjunct;
if(conjunct->dummy) return;
supList = conjunct->supList;
size = conjunct->nSize;
conjunct->dummy = cc_index;
for(i=0; i<size; i++) {
id = supList[i];
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] : 0;
if(!var) continue;
relArr = var->relArr;
for(j=0; j<var->nSize; j++) {
index = relArr[j];
if(index == MAXINT) continue;
if(index == MAXINT-1) continue;
tConjunct = head->conjunctArray[index];
if(tConjunct->dummy) continue;
ImgLinearFindConnectedComponent(head, tConjunct, cc_index);
}
}
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearFindSameSupportConjuncts | ( | Relation_t * | head, |
| int | from, | ||
| int | to | ||
| ) | [static] |
Function********************************************************************
Synopsis [Find the conjuncts that have same support variables.]
Description [Find the conjuncts that have same support variables.]
SideEffects []
SeeAlso []
Definition at line 4502 of file imgLinear.c.
{
Conjunct_t *con1, *con2;
int i, j;
for(i=from; i<=to; i++) {
con1 = head->conjunctArray[i];
if(con1 == 0) continue;
for(j=from; j<=to; j++) {
if(i == j) continue;
con2 = head->conjunctArray[j];
if(con2 == 0) continue;
if(ImgLinearIsSameConjunct(head, con1, con2)) {
ImgLinearAddConjunctIntoClusterArray(con1, con2);
head->conjunctArray[j] = 0;
head->bModified = 1;
head->bRefineVarArray = 1;
head->nTotalCluster++;
}
}
}
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearFreeSmoothArray | ( | array_t * | smoothVarBddArray | ) | [static] |
Function********************************************************************
Synopsis [Free smooth variable array]
Description [Free smooth variable array]
SideEffects []
SeeAlso []
Definition at line 3357 of file imgLinear.c.
{
int i;
array_t *smoothArray;
for(i=0; i<array_n(smoothVarBddArray); i++) {
smoothArray = array_fetch(array_t *, smoothVarBddArray, i);
mdd_array_free(smoothArray);
}
array_free(smoothVarBddArray);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| int* ImgLinearGetSupportBddId | ( | mdd_manager * | mgr, |
| bdd_t * | f, | ||
| int * | nSize | ||
| ) |
Function********************************************************************
Synopsis [Find support varaibles of bdd]
Description [Find support varaibles of bdd]
SideEffects []
SeeAlso []
Definition at line 487 of file imgLinear.c.
{
var_set_t *vset;
array_t *bvar_list;
int *supList, i, size;
bvar_list = mdd_ret_bvar_list(mgr);
vset = bdd_get_support(f);
size = 0;
supList = ALLOC(int, size+1);
supList[0] = -1;
for(i=0; i<array_n(bvar_list); i++) {
if(var_set_get_elt(vset, i) == 1) {
supList = REALLOC(int, supList, size+2);
supList[size++] = i;
supList[size] = -1;
}
}
var_set_free(vset);
*nSize = size;
if(size == 0) {
free(supList);
supList = 0;
}
return(supList);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearHeapCompareAffinityDeadLive | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3885 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = (Cluster_t *)c1;
clu2 = (Cluster_t *)c2;
if(clu1->nAffinity == clu2->nAffinity) {
if(clu1->nDead == clu2->nDead) {
return(clu1->nVar-clu1->nDead > clu2->nVar-clu2->nDead);
}
return(clu1->nDead < clu2->nDead);
}
return(clu1->nAffinity < clu2->nAffinity);
}
| static int ImgLinearHeapCompareDeadAffinityLive | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3831 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = (Cluster_t *)c1;
clu2 = (Cluster_t *)c2;
if(clu1->nDead == clu2->nDead) {
if(clu1->nAffinity == clu2->nAffinity) {
return((clu1->nVar-clu1->nDead) > (clu2->nVar-clu2->nDead));
}
return(clu1->nAffinity < clu2->nAffinity);
}
return(clu1->nDead < clu2->nDead);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearHeapCompareDeadLiveAffinity | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3858 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = (Cluster_t *)c1;
clu2 = (Cluster_t *)c2;
if(clu1->nDead == clu2->nDead) {
if((clu1->nVar-clu1->nDead) == (clu2->nVar-clu2->nDead)) {
return(clu1->nAffinity < clu2->nAffinity);
}
return((clu1->nVar-clu1->nDead) > (clu2->nVar-clu2->nDead));
}
return(clu1->nDead < clu2->nDead);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearHeapCompareLiveAffinityDead | ( | const void * | c1, |
| const void * | c2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Priority function for clutering]
Description [Priority function for clutering]
SideEffects []
SeeAlso []
Definition at line 3912 of file imgLinear.c.
{
Cluster_t *clu1, *clu2;
clu1 = (Cluster_t *)c1;
clu2 = (Cluster_t *)c2;
if(clu1->nVar-clu1->nDead == clu2->nVar-clu2->nDead) {
if(clu1->nAffinity == clu2->nAffinity) {
return(clu1->nDead < clu2->nDead);
}
return(clu1->nAffinity < clu2->nAffinity);
}
return(clu1->nVar-clu1->nDead > clu2->nVar-clu2->nDead);
}
| static void ImgLinearInsertClusterCandidate | ( | Relation_t * | head, |
| int | from, | ||
| int | to, | ||
| int | nDead, | ||
| int | nVar, | ||
| double | affinityLimit | ||
| ) | [static] |
Function********************************************************************
Synopsis [Add candidates for clustering]
Description [Add candidates for clustering, each condidate has pair of relation]
SideEffects []
SeeAlso []
if(affinity < affinityLimit) { st_free_table(table); return; }
Definition at line 1675 of file imgLinear.c.
{
Conjunct_t *conjunct;
Cluster_t *clu;
int *varType, *supList;
int i, j, id, size;
st_table *table;
double affinity, tAffinity;
int preSize, postSize, curSize, overlap;
varType = head->varType;
conjunct = head->conjunctArray[from];
if(conjunct->relation == 0) {
return;
}
supList = conjunct->supList;
size = conjunct->nSize;
table = st_init_table(st_numcmp, st_numhash);
for(i=0; i<size; i++) {
id = supList[i];
if(varType[id] == 2) continue;
st_insert(table, (char *)(long)id, (char *)(long)id);
}
tAffinity = 0.0;
for(i=from+1; i<=to; i++) {
preSize = table->num_entries;
conjunct = head->conjunctArray[i];
if(conjunct->relation == 0) continue;
supList = conjunct->supList;
size = conjunct->nSize;
curSize = 0;
for(j=0; j<size; j++) {
id = supList[j];
if(varType[id] == 2) continue;
curSize++;
st_insert(table, (char *)(long)id, (char *)(long)id);
}
postSize = table->num_entries;
overlap = curSize - (postSize-preSize);
if(curSize > preSize) affinity = (double)overlap/(double)preSize;
else affinity = (double)overlap/(double)curSize;
tAffinity += affinity;
}
st_free_table(table);
tAffinity /= (double)(to-from);
clu = ALLOC(Cluster_t, 1);
clu->from = from;
clu->to = to;
clu->nVar = nVar;
clu->nDead = nDead;
clu->nAffinity = (int)(tAffinity * 100.0);
head->clusterArray = REALLOC(Cluster_t *, head->clusterArray, head->nClusterArray+1);
head->clusterArray[head->nClusterArray++] = clu;
if(head->verbosity >= 5) {
fprintf(stdout, "Candidate Cluster : %4d-%4d G(%3d) V(%3d) A(%3d)\n",
clu->from, clu->to, clu->nDead, clu->nVar, clu->nAffinity);
fflush(stdout);
}
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearInsertPairClusterCandidate | ( | Relation_t * | head, |
| int | from, | ||
| double | affinityLimit, | ||
| int | varLimit, | ||
| int | rangeFlag | ||
| ) | [static] |
Function********************************************************************
Synopsis [Insert initial candidate set]
Description [Insert initial candidate set]
SideEffects []
SeeAlso []
Definition at line 2136 of file imgLinear.c.
{
Conjunct_t *conjunct, *fromC, *toC;
Cluster_t *clu;
int *varType, *supList;
st_table *table;
double affinity;
int preSize, postSize, curSize, overlap;
int i, j, id, tid, size, nDead;
int to, effTo;
VarLife_t *var;
if(from < 0) return;
if(from == head->nSize-1) return;
conjunct = head->conjunctArray[from];
if(conjunct == 0 || conjunct->relation == 0) {
for(; from>=0; from--) {
conjunct = head->conjunctArray[from];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
break;
}
}
if(from < 0) return;
to = -1;
for(i=from+1; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
if(!conjunct) continue;
if(conjunct->relation == 0) continue;
to = i;
break;
}
if(to == -1) return;
for(effTo = to+1; effTo<head->nSize; effTo++) {
conjunct = head->conjunctArray[effTo];
if(conjunct && conjunct->relation) break;
}
effTo--;
varType = head->varType;
fromC = head->conjunctArray[from];
toC = head->conjunctArray[to];
if(rangeFlag &&
(fromC->nSize == fromC->nRange || toC->nSize == toC->nRange))
return;
if(fromC->nSize == fromC->nRange && toC->nSize != toC->nRange)
return;
if(fromC->nSize != fromC->nRange && toC->nSize == toC->nRange)
return;
nDead = 0;
supList = fromC->supList;
size = fromC->nSize;
table = st_init_table(st_numcmp, st_numhash);
for(i=0; i<size; i++) {
id = supList[i];
if(varType[id] == 2) continue;
preSize++;
st_insert(table, (char *)(long)id, (char *)(long)id);
var = head->varArray[head->varArrayMap[id]];
if(from <= var->from && var->to <= effTo) nDead++;
}
preSize = table->num_entries;
supList = toC->supList;
size = toC->nSize;
curSize = 0;
for(j=0; j<size; j++) {
id = supList[j];
if(varType[id] == 2) continue;
curSize++;
if(st_lookup(table, (char *)(long)id, &tid)) continue;
st_insert(table, (char *)(long)id, (char *)(long)id);
var = head->varArray[head->varArrayMap[id]];
if(from <= var->from && var->to <= effTo) nDead++;
}
postSize = table->num_entries;
overlap = curSize - (postSize-preSize);
if(overlap == 0) affinity = 0.0;
else {
if(curSize > preSize)affinity = (double)overlap/(double)preSize;
else affinity = (double)overlap/(double)curSize;
}
st_free_table(table);
if(affinity < affinityLimit) return;
if(affinity == 0.0 && postSize > 10 &&
(fromC->bSingleton || toC->bSingleton)) return;
if(postSize-nDead > varLimit) return;
clu = ALLOC(Cluster_t, 1);
memset(clu, 0, sizeof(Cluster_t));
clu->from = from;
clu->to = to;
clu->nVar = postSize;
clu->nDead = nDead;
clu->nAffinity = (int)(affinity * 100.0);
Heap_HeapInsertCompare(head->clusterHeap, (void *)clu, (long)clu);
if(head->verbosity >= 5) {
fprintf(stdout, "Candidate Cluster : %4d -%4d G(%3d) V(%3d) A(%3d)\n",
clu->from, clu->to, clu->nDead, clu->nVar, clu->nAffinity);
fflush(stdout);
}
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearIsSameConjunct | ( | Relation_t * | head, |
| Conjunct_t * | con1, | ||
| Conjunct_t * | con2 | ||
| ) | [static] |
Function********************************************************************
Synopsis [Check if given conjuncts have same support variables]
Description [Check if given conjuncts have same support variables]
SideEffects []
SeeAlso []
Definition at line 4539 of file imgLinear.c.
{
int i, j;
int *sup1, *sup2;
int id1, id2;
int *varType;
if(con1->nDomain != con2->nDomain) return(0);
if(con1->nSize-con1->nRange != con2->nSize-con2->nRange) return(0);
sup1 = con1->supList;
sup2 = con2->supList;
varType = head->varType;
for(j=0, i=0; i<con1->nSize; i++) {
id1 = sup1[i];
if(varType[id1] == 2)continue;
while(1) {
id2 = sup2[j];
if(varType[id2] == 2) {
j++;
continue;
}
if(id1 != id2) return(0);
j++;
break;
}
}
return(1);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| array_t* ImgLinearMakeSmoothVarBdd | ( | Relation_t * | head, |
| bdd_t ** | smoothCubeArr | ||
| ) |
Function********************************************************************
Synopsis [Make the array of smooth variables]
Description [Make the array of smooth variables]
SideEffects []
SeeAlso []
Definition at line 3446 of file imgLinear.c.
{
array_t *smoothVarBddArray;
array_t *smoothArray;
VarLife_t **varArray;
int i, j, id;
VarLife_t *var;
bdd_t *varBdd, *cube, *newCube;
smoothVarBddArray = array_alloc(array_t *, 0);
for(i=0; i<=head->nSize; i++) {
smoothArray = array_alloc(bdd_t *, 0);
array_insert_last(array_t *, smoothVarBddArray, smoothArray);
}
varArray = head->varArray;
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(var->stateVar == 2) continue;
smoothArray = array_fetch(array_t *, smoothVarBddArray, var->to+1);
varBdd = bdd_get_variable(head->mgr, var->id);
array_insert_last(bdd_t *, smoothArray, varBdd);
}
smoothArray = array_fetch(array_t *, smoothVarBddArray, 0);
for(i=0; i<head->nDomain; i++) {
id = head->domainVars[i];
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] : 0;
if(var) continue;
varBdd = bdd_get_variable(head->mgr, id);
array_insert_last(bdd_t *, smoothArray, varBdd);
}
if(smoothCubeArr) {
for(i=0; i<=head->nSize; i++) {
smoothArray = array_fetch(array_t *, smoothVarBddArray, i);
cube = bdd_one(head->mgr);
for(j=0; j<array_n(smoothArray); j++) {
varBdd = array_fetch(bdd_t *, smoothArray, j);
newCube = bdd_and(varBdd, cube, 1, 1);
bdd_free(cube);
cube = newCube;
}
smoothCubeArr[i] = cube;
}
}
return(smoothVarBddArray);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| int ImgLinearOptimizeAll | ( | Relation_t * | head, |
| Img_OptimizeType | optDir, | ||
| int | constantNSOpt | ||
| ) |
Function********************************************************************
Synopsis [Function for optimizing state varaibles]
Description [If the next or current state variables are eleminated after applying clustering and constant propagation then delete corresponding state variable from transition relation.]
SideEffects []
SeeAlso []
Definition at line 581 of file imgLinear.c.
{
int bOptimize;
bOptimize = 0;
bOptimize |= ImgLinearPropagateConstant(head, constantNSOpt);
bOptimize |= ImgLinearOptimizeStateVariables(head, optDir);
ImgLinearOptimizeInternalVariables(head);
return(bOptimize);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearOptimizeInternalVariables | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Remove intermediate variables after propagating constant.]
Description [Remove intermediate variables after propagating constant.]
SideEffects []
SeeAlso []
Definition at line 836 of file imgLinear.c.
{
bdd_t *oneBdd, *varBdd;
VarLife_t **varArray, *var;
Conjunct_t **conjunctArray;
array_t *smoothVarBddArray;
int *varType;
int i, bModified;
oneBdd = bdd_one(head->mgr);
while(1) {
bModified = 0;
varArray = head->varArray;
varType = head->varType;
conjunctArray = head->conjunctArray;
for(i=0; i<head->nVarArray; i++) {
var = head->varArray[i];
if(varType[var->id] == 2)
continue;
if(varType[var->id] == 1 && head->quantifyCS == 0)
continue;
if(var->from == var->to) {
if(conjunctArray[var->from] == 0) continue;
varBdd = bdd_get_variable(head->mgr, var->id);
smoothVarBddArray = array_alloc(bdd_t *, 0);
array_insert_last(bdd_t *, smoothVarBddArray, varBdd);
ImgLinearQuantifyVariablesFromConjunct(
head, head->conjunctArray[var->from], smoothVarBddArray, &bModified);
mdd_array_free(smoothVarBddArray);
}
}
if(bModified == 0) break;
ImgLinearVariableArrayInit(head);
}
bdd_free(oneBdd);
return;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| int ImgLinearOptimizeStateVariables | ( | Relation_t * | head, |
| Img_OptimizeType | optDir | ||
| ) |
Function********************************************************************
Synopsis [Remove corresponding current (next) state variables if the next (current state variables are eleminated aftetr clustering]
Description [Remove corresponding current (next) state variables if the next (current state variables are eleminated aftetr clustering]
SideEffects []
SeeAlso []
Definition at line 706 of file imgLinear.c.
{
int *rangeId, *domainId, *existStateVar;
int *domain2range;
int *range2domain;
bdd_t *oneBdd, *varBdd;
VarLife_t **varArray, *var;
array_t *smoothVarBddArray;
Conjunct_t *conjunct;
int i, id;
int bOptimize;
int rangeBound, domainBound;
int nRangeReduced, nDomainReduced;
if(optDir == Opt_None) return(0);
id = 0;
ImgLinearVariableArrayInit(head);
bOptimize = 0;
rangeId = ALLOC(int, head->nRange+1);
memset(rangeId, 0, sizeof(int)*(head->nRange+1));
domainId = ALLOC(int, head->nDomain+1);
memset(domainId, 0, sizeof(int)*(head->nDomain+1));
existStateVar = ALLOC(int, head->nVar);
memset(existStateVar, 0, sizeof(int)*head->nVar);
ImgLinearSetEffectiveNumberOfStateVariable(head, rangeId, domainId, existStateVar);
rangeBound = (int)((double)head->nInitRange * BACKWARD_REDUCTION_RATE);
domainBound = (int)((double)head->nInitDomain * FORWARD_REDUCTION_RATE);
if(head->nEffDomain < domainBound) {
if(optDir == Opt_Both) optDir = Opt_NS;
else if(optDir == Opt_CS) optDir = Opt_None;
}
if(head->nEffRange < rangeBound) {
if(optDir == Opt_Both) optDir = Opt_CS;
else if(optDir == Opt_NS) optDir = Opt_None;
}
if(optDir == Opt_None) return(0);
oneBdd = bdd_one(head->mgr);
domain2range = head->domain2range;
range2domain = head->range2domain;
varArray = head->varArray;
if(optDir == Opt_Both || optDir == Opt_CS) {
smoothVarBddArray = array_alloc(bdd_t *, 0);
nDomainReduced = 0;
for(i=0; i<head->nEffDomain; i++) {
id = domainId[i];
if(existStateVar[domain2range[id]]) continue;
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] : 0;
if(var) {
if(head->nEffDomain-nDomainReduced < domainBound) break;
if(var->nSize-head->includeCS == 1) {
varBdd = bdd_get_variable(head->mgr, id);
array_insert_last(bdd_t *, smoothVarBddArray, varBdd);
nDomainReduced++;
}
}
}
if(array_n(smoothVarBddArray)) {
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
if(conjunct->nDomain == 0) continue;
if(ImgLinearQuantifyVariablesFromConjunct(
head, conjunct, smoothVarBddArray, &bOptimize)) {
fprintf(vis_stdout, "NOTICE : CS %3d quantified from %d'th relation\n", id, i);
}
}
}
mdd_array_free(smoothVarBddArray);
}
if(optDir == Opt_Both || optDir == Opt_NS) {
smoothVarBddArray = array_alloc(bdd_t *, 0);
nRangeReduced = 0;
for(i=0; i<head->nEffRange; i++) {
id = rangeId[i];
if(existStateVar[range2domain[id]]) continue;
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] :
0;
if(var) {
if(head->nEffRange-nRangeReduced < rangeBound) break;
if(var->nSize-head->includeNS == 1) {
varBdd = bdd_get_variable(head->mgr, id);
array_insert_last(bdd_t *, smoothVarBddArray, varBdd);
nRangeReduced++;
}
}
}
if(array_n(smoothVarBddArray)) {
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
if(conjunct->nRange == 0) continue;
if(ImgLinearQuantifyVariablesFromConjunct(
head, conjunct, smoothVarBddArray, &bOptimize)) {
fprintf(vis_stdout, "NOTICE : NS %3d quantified from %d'th relation\n", id, i);
}
}
}
mdd_array_free(smoothVarBddArray);
}
bdd_free(oneBdd);
free(rangeId);
free(domainId);
free(existStateVar);
ImgLinearVariableArrayInit(head);
return(bOptimize);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearPrintDebugInfo | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Print information of debug information]
Description [Print information of debug information]
SideEffects []
SeeAlso []
Definition at line 2266 of file imgLinear.c.
{
int i, j, id;
Conjunct_t *conjunct;
int *supList;
array_t *smoothVarBddArray;
array_t *smoothArray;
bdd_t *varBdd;
int idPerLine;
idPerLine = 12;
fprintf(vis_stdout, "-----------------------------------------------------\n");
fprintf(vis_stdout, " Debug Information for Transition Relations\n");
fprintf(vis_stdout, "-----------------------------------------------------\n");
fprintf(vis_stdout, "List of Quantify Variables---------------------------\n");
for(i=0; i<head->nQuantify; i++) {
if(i!=0 && i%idPerLine == 0) fprintf(vis_stdout, "\n");
fprintf(vis_stdout, "%3d ", head->quantifyVars[i]);
}
fprintf(vis_stdout, "\n");
fprintf(vis_stdout, "List of Domain Variables-----------------------------\n");
for(i=0; i<head->nDomain; i++) {
if(i!=0 && i%idPerLine == 0) fprintf(vis_stdout, "\n");
fprintf(vis_stdout, "%3d ", head->domainVars[i]);
}
fprintf(vis_stdout, "\n");
fprintf(vis_stdout, "List of Range Variables------------------------------\n");
for(i=0; i<head->nRange; i++) {
if(i!=0 && i%idPerLine == 0) fprintf(vis_stdout, "\n");
fprintf(vis_stdout, "%3d ", head->rangeVars[i]);
}
fprintf(vis_stdout, "\n");
fprintf(vis_stdout, "Varibles in Each Transition Relation-----------------\n");
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
supList = conjunct->supList;
fprintf(vis_stdout, "%3d'th : ", i);
for(j=0; j<conjunct->nSize; j++) {
if(j!=0 && j%idPerLine == 0) fprintf(vis_stdout, "\n ");
fprintf(vis_stdout, "%3d ", supList[j]);
}
fprintf(vis_stdout, "\n");
}
smoothVarBddArray = ImgLinearMakeSmoothVarBdd(head, 0);
fprintf(vis_stdout, "Quantified Schedule----------------------------------\n");
for(i=0; i<=head->nSize; i++) {
fprintf(vis_stdout, "%3d'th : ", i);
smoothArray = array_fetch(array_t *, smoothVarBddArray, i);
for(j=0; j<array_n(smoothArray); j++) {
varBdd = array_fetch(bdd_t *, smoothArray, j);
id = bdd_top_var_id(varBdd);
if(j!=0 && j%idPerLine == 0) fprintf(vis_stdout, "\n ");
fprintf(vis_stdout, "%3d ", id);
}
fprintf(vis_stdout, "\n");
}
ImgLinearFreeSmoothArray(smoothVarBddArray);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearPrintMatrix | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Print matrix]
Description [Print matrix. Its x axis is varaible and its y axis is relation]
SideEffects [Print matrix. Its x axis is varaible and its y axis is relation]
SeeAlso []
Definition at line 3118 of file imgLinear.c.
{
int i, j, id, index;
int *mapArray;
VarLife_t **varArray, *var;
VarLife_t **auxArr, **sortArr;
int nAuxArr, nSortArr;
array_t *smoothVarBddArr, *smoothArray;
bdd_t *varBdd;
Conjunct_t *conjunct;
bdd_t *relation;
int *supList, *varType;
char *buffer;
int maxIndex;
if(head->verbosity < 5) return;
ImgLinearVariableArrayInit(head);
mapArray = ALLOC(int, head->nVar);
memset(mapArray, -1, sizeof(int)*head->nVar);
varArray = head->varArray;
smoothVarBddArr = ImgLinearMakeSmoothVarBdd(head, 0);
for(index=0, i=0; i<array_n(smoothVarBddArr); i++) {
smoothArray = array_fetch(array_t *, smoothVarBddArr, i);
nAuxArr = 0;
auxArr = ALLOC(VarLife_t *, nAuxArr+1);
for(j=0; j<array_n(smoothArray); j++) {
varBdd = array_fetch(bdd_t *, smoothArray, j);
id = (int)bdd_top_var_id(varBdd);
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] :
0;
if(var) {
auxArr[nAuxArr++] = var;
auxArr = REALLOC(VarLife_t *, auxArr, nAuxArr+1);
}
}
qsort(auxArr, nAuxArr, sizeof(VarLife_t *), ImgLinearCompareVarIndex);
for(j=0; j<nAuxArr; j++)
mapArray[auxArr[j]->id] = index++;
free(auxArr);
}
varArray = head->varArray;
nSortArr = 0;
sortArr = ALLOC(VarLife_t *, nSortArr+1);
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(var->stateVar == 2) continue;
if(mapArray[var->id] >= 0) {
var->index = mapArray[var->id];
sortArr[nSortArr++] = var;
sortArr = REALLOC(VarLife_t *, sortArr, nSortArr+1);
}
else {
var->index = (double)MAXINT;
}
}
qsort(sortArr, nSortArr, sizeof(VarLife_t *), ImgLinearCompareVarIndex);
fprintf(stdout, " ");
for(i=0; i<nSortArr; i++) fprintf(stdout, "%d", (i/10)%10);
fprintf(stdout, "\n");
fprintf(stdout, " ");
for(i=0; i<nSortArr; i++) fprintf(stdout, "%d", i%10);
fprintf(stdout, "\n");
buffer = ALLOC(char , nSortArr+1);
for(i=0; i<nSortArr; i++) {
var = sortArr[i];
var->index = (double)i;
if(var->stateVar == 1) buffer[i] = 'C';
else buffer[i] = 'T';
}
buffer[i] = '\0';
free(sortArr);
fprintf(stdout, " %s\n", buffer);
maxIndex = i;
for(i=0; i<maxIndex; i++) buffer[i] = '.';
buffer[i] = '\0';
varType = head->varType;
for(index=0,i=head->nSize-1;i>=0; i--) {
conjunct = head->conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
relation = conjunct->relation;
if(relation == (mdd_t *)(MAXINT-1)) continue;
if(relation == 0) continue;
supList = conjunct->supList;
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
if(varType[id] == 2) continue;
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] :
0;
if(!var) continue;
if((int)var->index == MAXINT) continue;
buffer[(int)var->index] = '1';
}
index++;
fprintf(stdout, "%4d %s %d %d\n", i, buffer, conjunct->nRange, bdd_size(relation));
fflush(stdout);
for(j=0; j<maxIndex; j++) buffer[j] = '.';
buffer[j] = '\0';
}
free(buffer);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearPrintMatrixFull | ( | Relation_t * | head, |
| int | matrixIndex | ||
| ) | [static] |
Function********************************************************************
Synopsis [Print matrix]
Description [Print matrix]
SideEffects []
SeeAlso []
Definition at line 3242 of file imgLinear.c.
{
int i, j, id, index;
int *mapArray;
VarLife_t **varArray, *var;
VarLife_t **auxArr, **sortArr;
int nAuxArr, nSortArr;
array_t *smoothVarBddArr, *smoothArray;
FILE *fout;
int min, max;
bdd_t *varBdd;
char filename[1024];
Conjunct_t *conjunct;
bdd_t *relation;
int *supList;
ImgLinearVariableArrayInit(head);
mapArray = ALLOC(int, head->nVar);
memset(mapArray, -1, sizeof(int)*head->nVar);
varArray = head->varArray;
smoothVarBddArr = ImgLinearMakeSmoothVarBdd(head, 0);
for(index=0, i=0; i<array_n(smoothVarBddArr); i++) {
smoothArray = array_fetch(array_t *, smoothVarBddArr, i);
nAuxArr = 0;
auxArr = ALLOC(VarLife_t *, nAuxArr+1);
for(j=0; j<array_n(smoothArray); j++) {
varBdd = array_fetch(bdd_t *, smoothArray, j);
id = (int)bdd_top_var_id(varBdd);
var = head->varArrayMap[id] >=0 ? varArray[head->varArrayMap[id]] : 0;
if(var) {
auxArr[nAuxArr++] = var;
auxArr = REALLOC(VarLife_t *, auxArr, nAuxArr+1);
}
}
qsort(auxArr, nAuxArr, sizeof(VarLife_t *), ImgLinearCompareVarIndex);
for(j=0; j<nAuxArr; j++)
mapArray[auxArr[j]->id] = index++;
free(auxArr);
}
nSortArr = 0;
sortArr = ALLOC(VarLife_t *, nSortArr+1);
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(var->stateVar == 2) continue;
if(mapArray[var->id] >= 0) {
var->index = mapArray[var->id];
sortArr[nSortArr++] = var;
sortArr = REALLOC(VarLife_t *, sortArr, nSortArr+1);
}
else {
var->index = (double)MAXINT;
}
}
qsort(sortArr, nSortArr, sizeof(VarLife_t *), ImgLinearCompareVarIndex);
free(mapArray);
sprintf(filename, "fullmatrix%d.data", matrixIndex);
fout = fopen(filename, "w");
min = 0;
max = head->nSize+1;
for(i=0; i<nSortArr; i++) {
var = sortArr[i];
if(var->stateVar == 1)
fprintf(fout, "%d %d\n", (int)var->index, min);
}
free(sortArr);
for(index=0, i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
relation = conjunct->relation;
if(relation == (mdd_t *)(MAXINT-1)) continue;
if(relation == 0) continue;
supList = conjunct->supList;
for(j=0; j<conjunct->nSize; j++) {
id = supList[j];
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] : 0;
if(!var) continue;
if(var->stateVar == 2)
if((int)var->index == MAXINT) continue;
fprintf(fout, "%d %d\n", (int)var->index, index+1);
}
index++;
}
fclose(fout);
sprintf(filename, "fullmatrix%d.gnu", matrixIndex);
fout = fopen(filename, "w");
fprintf(fout, "set terminal postscript \n");
fprintf(fout, "set output \"fullmatrix%d.ps\"\n", matrixIndex);
fprintf(fout, "set title \"profile of live variable\"\n");
fprintf(fout, "set yrange [-1:%d]\n", head->nSize+2);
fprintf(fout, "plot \"fullmatrix%d.data\" using 1:2 title \"%d\" with point\n",
matrixIndex, matrixIndex);
fclose(fout);
ImgLinearFreeSmoothArray(smoothVarBddArr);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearPrintTransitionInfo | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Print transition relation info]
Description [Print transition relation info]
SideEffects []
SeeAlso []
Definition at line 3415 of file imgLinear.c.
{
double aal, atl;
fprintf(stdout, "SUMMARY : domain variables %d -> %d -> %d\n",
head->nDomain, head->nInitDomain, head->nEffDomain);
fprintf(stdout, "SUMMARY : range variables %d -> %d -> %d\n",
head->nRange, head->nInitRange, head->nEffRange);
fprintf(stdout, "SUMMARY : quantify variables %d -> %d -> %d\n",
head->nQuantify, head->nInitQuantify, head->nEffQuantify);
fprintf(stdout, "SUMMARY : number of transition relation %d\n",
head->nSize);
ImgLinearComputeLifeTime(head, &aal, &atl);
fprintf(stdout, "Active Life Time : %g\n", aal);
fprintf(stdout, "Average Life Time : %g\n", atl);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearPrintVariableProfile | ( | Relation_t * | head, |
| char * | baseName | ||
| ) | [static] |
Function********************************************************************
Synopsis [Print profile of variables]
Description [Print profile of variables]
SideEffects []
SeeAlso []
Definition at line 3045 of file imgLinear.c.
{
double aal, atl;
char filename[1024];
FILE *fout;
st_table *cutSet;
int i, j, nDead, id;
int *varType;
int *supList;
Conjunct_t *conjunct;
VarLife_t *var;
st_generator *gen;
ImgLinearVariableArrayInit(head);
varType = head->varType;
sprintf(filename, "%s.data", baseName);
fout = fopen(filename, "w");
cutSet = st_init_table(st_numcmp, st_numhash);
for(i=0; i<head->nVar; i++) {
if(varType[i] == 1)
st_insert(cutSet, (char *)(long)i, (char *)(long)i);
}
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
supList = conjunct->supList;
for(j=0; j<conjunct->nSize; j++)
st_insert(cutSet, (char *)(long)supList[j], (char *)(long)supList[j]);
nDead = 0;
st_foreach_item(cutSet, gen, &id, &id) {
if(varType[id] == 2) continue;
var = head->varArrayMap[id] >=0 ? head->varArray[head->varArrayMap[id]] :
0;
if(!var) continue;
if(var->to <= i) {
nDead++;
st_delete(cutSet, (&id), NULL);
}
}
fprintf(fout, "%d %d %d\n", i, cutSet->num_entries, cutSet->num_entries+nDead);
}
fclose(fout);
ImgLinearComputeLifeTime(head, &aal, &atl);
sprintf(filename, "%s.gnu", baseName);
fout = fopen(filename, "w");
fprintf(fout, "set terminal postscript \n");
fprintf(fout, "set output \"%s.ps\"\n", baseName);
fprintf(fout, "set title \"profile of live variable aal %.3f atl %.3f\"\n",
aal, atl);
fprintf(fout, "plot \"%s.data\" using 1:2 title \"%s\" with lines, \\\n", baseName, "cut");
fprintf(fout, " \"%s.data\" using 1:3 title \"%s\" with lines\n", baseName, "cut+dead");
fclose(fout);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| int ImgLinearPropagateConstant | ( | Relation_t * | head, |
| int | nextStateFlag | ||
| ) |
Function********************************************************************
Synopsis [Propagate the constant to simply the transition relation]
Description [Propagate the constant to simply the transition relation]
SideEffects []
SeeAlso []
Definition at line 603 of file imgLinear.c.
{
int nSize, i, j;
int *supList;
Conjunct_t **conjunctArray;
Conjunct_t *conjunct;
mdd_manager *mgr;
bdd_t **constantCase;
bdd_t *relation, *simpleRelation;
int nConstantCase;
int bOptimize;
int id, cid, index;
int *varType;
int size;
bOptimize = 0;
mgr = head->mgr;
constantCase = head->constantCase;
varType = head->varType;
conjunctArray = head->conjunctArray;
nSize = head->nSize;
nConstantCase = 0;
for(index=0, i=0; i<nSize; i++) {
conjunct = conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
if(conjunct->nSize == 1) {
id = conjunct->supList[0];
if(varType[id] == 2) {
if(nextStateFlag) {
cid = head->range2domain[id];
if(constantCase[cid] == 0) {
constantCase[cid] = bdd_dup(conjunct->relation);
if(head->verbosity >= 4)
fprintf(vis_stdout, "Detect Constant Case %3d(%c)\n", cid, linearVarString[varType[cid]]);
nConstantCase++;
}
}
continue;
}
else {
if(constantCase[id] == 0) {
constantCase[id] = bdd_dup(conjunct->relation);
if(head->verbosity >= 4)
fprintf(vis_stdout, "Detect Constant Case %3d(%c)\n", id, linearVarString[varType[id]]);
nConstantCase++;
}
continue;
}
}
}
if(nConstantCase > 0) {
for(i=0; i<head->nSize; i++) {
conjunct = conjunctArray[i];
if(conjunct == 0) continue;
if(conjunct->relation == 0) continue;
relation = conjunct->relation;
size = conjunct->nSize;
supList = conjunct->supList;
for(j=0; j<size; j++) {
id = supList[j];
if(constantCase[id] == 0) continue;
simpleRelation = bdd_cofactor(relation, constantCase[id]);
if(bdd_equal(relation, simpleRelation)) {
bdd_free(simpleRelation);
continue;
}
if(varType[id] == 1) bOptimize++;
bdd_free(conjunct->relation);
conjunct->relation = simpleRelation;
relation = conjunct->relation;
conjunct->bModified = 1;
head->bModified = 1;
head->bRefineVarArray = 1;
if(head->verbosity >= 2)
fprintf(vis_stdout, "Constant propagation is applied %3d(%c) to %3d'th relation\n", id, linearVarString[varType[id]], i);
}
ImgLinearConjunctRefine(head, conjunct);
}
}
return(bOptimize);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static int ImgLinearQuantifyVariablesFromConjunct | ( | Relation_t * | head, |
| Conjunct_t * | conjunct, | ||
| array_t * | smoothVarBddArray, | ||
| int * | bModified | ||
| ) | [static] |
Function********************************************************************
Synopsis [Apply quantification with candidate variables]
Description [Apply quantification with candidate variables]
SideEffects []
SeeAlso []
Definition at line 4104 of file imgLinear.c.
{
bdd_t *relation, *simpleRelation;
if(conjunct == 0) return 1;
relation = conjunct->relation;
if(relation == 0) return 1;
simpleRelation = bdd_smooth(relation, smoothVarBddArray);
if(bdd_equal(relation, simpleRelation)) {
bdd_free(simpleRelation);
return 0;
}
bdd_free(relation);
(*bModified)++;
conjunct->relation = simpleRelation;
conjunct->bModified = 1;
head->bModified = 1;
head->bRefineVarArray = 1;
ImgLinearConjunctRefine(head, conjunct);
return 1;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearRefineRelation | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Refine transition relation.]
Description [Clean up the Relation_t data structure after applying linear arrangement of transition relation]
SideEffects []
SeeAlso []
Definition at line 889 of file imgLinear.c.
{
ImgLinearConjunctArrayRefine(head);
ImgLinearVariableArrayInit(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| Relation_t* ImgLinearRelationInit | ( | mdd_manager * | mgr, |
| array_t * | relationArray, | ||
| array_t * | domainBddVars, | ||
| array_t * | rangeBddVars, | ||
| array_t * | quantifyBddVars, | ||
| ImgTrmOption_t * | option | ||
| ) |
Function********************************************************************
Synopsis [Initialize the data structure for linear arrangement and clustering.]
Description [Build initial data structure]
SideEffects []
SeeAlso []
Definition at line 330 of file imgLinear.c.
{
Relation_t *head;
Conjunct_t *conjunct, **conjunctArray;
bdd_t *varBdd;
int *domain2range, *range2domain;
int *varArrayMap, *varType;
bdd_t **constantCase, *relation;
int i, j, id, tid, nSize;
int *supList, listSize;
char *flagValue;
int *quantifyVars, nQuantify, *visited;
head = ALLOC(Relation_t, 1);
memset(head, 0, sizeof(Relation_t));
head->mgr = mgr;
head->nVar = bdd_num_vars(head->mgr);
head->bddLimit = option->clusterSize;
head->verbosity = option->verbosity;
flagValue = Cmd_FlagReadByName("linear_verbosity");
if(flagValue) {
head->verbosity += atoi(flagValue);
}
head->includeCS = option->linearIncludeCS;
head->includeNS = option->linearIncludeNS;
head->quantifyCS = option->linearQuantifyCS;
head->computeRange = option->linearComputeRange;
head->nInitRange = 0;
head->nInitDomain = 0;
head->nInitQuantify = 0;
head->nEffRange = 0;
head->nEffDomain = 0;
head->nEffQuantify = 0;
head->clusterArray = 0;
head->nClusterArray = 0;
head->nVarArray = 0;
head->varArray = 0;
head->nSingletonArray = 0;
head->singletonArray = 0;
head->nNextStateArray = 0;
head->nextStateArray = 0;
domain2range = ALLOC(int, head->nVar);
head->domain2range = domain2range;
memset(domain2range, -1, sizeof(int)*head->nVar);
range2domain = ALLOC(int, head->nVar);
head->range2domain = range2domain;
memset(range2domain, -1, sizeof(int)*head->nVar);
varType = ALLOC(int, head->nVar);
head->varType = varType;
memset(varType, 0, sizeof(int)*head->nVar);
head->nRange = array_n(rangeBddVars);
head->nDomain = array_n(domainBddVars);
head->domainVars = ALLOC(int, head->nDomain);
head->rangeVars = ALLOC(int, head->nRange);
for(i=0; i<head->nDomain; i++) {
varBdd = array_fetch(bdd_t *, domainBddVars, i);
id = (int)bdd_top_var_id(varBdd);
head->domainVars[i] = id;
varType[id] = 1;
varBdd = array_fetch(bdd_t *, rangeBddVars, i);
tid = (int)bdd_top_var_id(varBdd);
head->rangeVars[i] = tid;
varType[tid] = 2;
domain2range[id] = tid;
range2domain[tid] = id;
}
head->quantifyVars = quantifyVars = ALLOC(int, head->nVar);
memset(quantifyVars, 0, sizeof(int)*head->nVar);
for(nQuantify=0, i=0; i<array_n(quantifyBddVars); i++) {
varBdd = array_fetch(bdd_t *, quantifyBddVars, i);
id = (int)bdd_top_var_id(varBdd);
quantifyVars[nQuantify++] = id;
varType[id] = 3;
}
varArrayMap = ALLOC(int, head->nVar);
head->varArrayMap = varArrayMap;
memset(varArrayMap, -1, sizeof(int)*head->nVar);
constantCase = ALLOC(bdd_t *, head->nVar);
head->constantCase = constantCase;
memset(constantCase, 0, sizeof(bdd_t *)*head->nVar);
nSize = array_n(relationArray);
conjunctArray = ALLOC(Conjunct_t *, nSize+1);
head->nSize = nSize;
head->conjunctArray = conjunctArray;
visited = ALLOC(int, head->nVar);
memset(visited, 0, sizeof(int)*head->nVar);
for(i=0; i<nSize; i++) {
conjunct = ALLOC(Conjunct_t, 1);
memset(conjunct, 0, sizeof(Conjunct_t));
conjunct->index = i;
relation = array_fetch(bdd_t *, relationArray, i);
conjunct->relation = mdd_dup(relation);
supList = ImgLinearGetSupportBddId(head->mgr, relation, &listSize);
conjunct->supList = supList;
conjunct->nSize = listSize;
for(j=0; j<listSize; j++) {
id = supList[j];
if(varType[id] == 1) conjunct->nDomain++;
else if(varType[id] == 2) conjunct->nRange++;
else if(varType[id] == 3) conjunct->nQuantify++;
else if(visited[id] == 0) {
quantifyVars[nQuantify++] = id;
visited[id] = 1;
}
}
conjunctArray[i] = conjunct;
}
conjunctArray[i] = 0;
head->nQuantify= nQuantify;
free(visited);
ImgLinearVariableArrayInit(head);
return(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| void ImgLinearRelationQuit | ( | Relation_t * | head | ) |
Function********************************************************************
Synopsis [Free Relation_t data structure]
Description [Free Relation_t data structure]
SideEffects []
SeeAlso []
Definition at line 1389 of file imgLinear.c.
{
int i;
Conjunct_t *conjunct;
if(head->varArray) ImgLinearVariableArrayQuit(head);
if(head->rangeVars) free(head->rangeVars);
if(head->domainVars) free(head->domainVars);
if(head->quantifyVars) free(head->quantifyVars);
if(head->varType) free(head->varType);
if(head->varArrayMap) free(head->varArrayMap);
if(head->singletonArray) free(head->singletonArray);
if(head->nextStateArray) free(head->nextStateArray);
if(head->constantCase) {
for(i=0; i<head->nVar; i++)
if(head->constantCase[i]) bdd_free(head->constantCase[i]);
}
for(i=0; i<head->nSize; i++) {
conjunct = head->conjunctArray[i];
ImgLinearConjunctQuit(conjunct);
}
free(head);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearSetEffectiveNumberOfStateVariable | ( | Relation_t * | head, |
| int * | rangeId, | ||
| int * | domainId, | ||
| int * | existStateVariable | ||
| ) | [static] |
Function********************************************************************
Synopsis [Get the number of state variables after applying optimization]
Description [Get the number of state variables after applying optimization]
SideEffects []
SeeAlso []
Definition at line 4260 of file imgLinear.c.
{
VarLife_t **varArray, *var;
int *varType;
int nRange, nDomain, nQuantify;
int i;
varArray = head->varArray;
varType = head->varType;
nRange = nDomain = nQuantify = 0;
for(i=0; i<head->nVarArray; i++) {
var = varArray[i];
if(varType[var->id] == 1) {
if(domainId) domainId[nDomain] = var->id;
nDomain++;
if(existStateVariable) existStateVariable[var->id] = 1;
}
else if(varType[var->id] == 2) {
if(rangeId) rangeId[nRange] = var->id;
nRange++;
if(existStateVariable) existStateVariable[var->id] = 1;
}
else if(varType[var->id] == 3) {
nQuantify++;
}
else
nQuantify++;
}
if(domainId) domainId[nDomain] = -1;
if(rangeId) rangeId[nRange] = -1;
head->nEffRange = nRange;
head->nEffDomain = nDomain;
head->nEffQuantify = nQuantify;
if(head->nInitRange == 0 && head->nInitDomain == 0 && head->nInitQuantify == 0) {
head->nInitRange = nRange;
head->nInitDomain = nDomain;
head->nInitQuantify = nQuantify;
}
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearUpdateVariableArrayWithId | ( | Relation_t * | head, |
| int | cindex, | ||
| int | id | ||
| ) | [static] |
AutomaticStart
Function********************************************************************
Synopsis [Update variable array with id of variable id]
Description [Update variable array with id of variable id]
SideEffects []
SeeAlso []
Definition at line 4030 of file imgLinear.c.
{
VarLife_t *var;
int *varType;
int *varArrayMap;
int nVarArray;
VarLife_t **varArray;
varType = head->varType;
varArrayMap = head->varArrayMap;
varArray = head->varArray;
nVarArray = head->nVarArray;
if(varArrayMap[id] == -1) {
var = ALLOC(VarLife_t, 1);
var->id = id;
var->from = cindex;
var->to = cindex;
var->effFrom = cindex;
var->effTo = cindex;
var->stateVar = 0;
var->relArr = ALLOC(int, 1);
var->nSize = 0;
var->index = 0.0;
varArrayMap[id] = nVarArray;
varArray[nVarArray++] = var;
varArray = REALLOC(VarLife_t *, varArray, nVarArray+1);
if(varType[id] == 1) {
var->stateVar = 1;
if(head->includeCS && !head->quantifyCS) {
var->relArr = REALLOC(int, var->relArr, var->nSize+1);
var->relArr[var->nSize++] = MAXINT-1;
var->from = -1;
}
}
else if(varType[id] == 2) {
var->stateVar = 2;
if(head->includeNS) {
var->relArr = REALLOC(int, var->relArr, var->nSize+1);
var->relArr[var->nSize++] = MAXINT;
var->to = head->nSize;
}
}
}
else {
var = varArray[varArrayMap[id]];
}
if(var->to < cindex) var->to = cindex;
if(var->from > cindex) var->from = cindex;
if(var->effTo < cindex) var->effTo = cindex;
if(var->effFrom > cindex) var->effFrom = cindex;
var->relArr = REALLOC(int, var->relArr, var->nSize+1);
var->relArr[var->nSize++] = cindex;
head->varArray = varArray;
head->nVarArray = nVarArray;
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearVariableArrayInit | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Initialize variable array]
Description [Initialize variable array]
SideEffects []
SeeAlso []
Definition at line 3939 of file imgLinear.c.
{
mdd_manager *mgr;
Conjunct_t **conjunctArray, *conjunct;
Conjunct_t *tConjunct;
bdd_t *relation, *bddOne;
VarLife_t **varArray;
int *supList, listSize;
int *varType, *varArrayMap;
int i, j, k, id, nVarArray;
int nSize, newNum;
if(head->bRefineVarArray) {
ImgLinearVariableArrayQuit(head);
}
if(head->varArray) return;
mgr = head->mgr;
if(head->nVar != (int)(bdd_num_vars(mgr))) {
newNum = bdd_num_vars(head->mgr);
head->domain2range = REALLOC(int, head->domain2range, newNum);
head->range2domain = REALLOC(int, head->range2domain, newNum);
head->varType = REALLOC(int, head->varType, newNum);
head->varArrayMap = REALLOC(int, head->varArrayMap, newNum);
head->constantCase = REALLOC(bdd_t *, head->constantCase, newNum);
head->quantifyVars = REALLOC(int, head->quantifyVars, newNum);
for(i=head->nVar; i<newNum; i++) {
head->domain2range[i] = -1;
head->range2domain[i] = -1;
head->varType[i] = 0;
head->varArrayMap[i] = -1;
head->constantCase[i] = 0;
head->quantifyVars[i] = 0;
}
head->nVar = bdd_num_vars(head->mgr);
}
nSize = head->nSize;
conjunctArray = head->conjunctArray;
varArrayMap = head->varArrayMap;
id = 0;
nVarArray = 0;
varArray = ALLOC(VarLife_t *, nVarArray+1);
head->varArray = varArray;
head->nVarArray = nVarArray;
bddOne = bdd_one(head->mgr);
varType = head->varType;
for(i=0; i<nSize; i++) {
conjunct = conjunctArray[i];
if(conjunct == 0) continue;
relation = conjunct->relation;
if(relation == 0) continue;
if(bdd_equal(bddOne, relation)) continue;
supList = conjunct->supList;
listSize = conjunct->nSize;
for(j=0; j<listSize; j++) {
id = supList[j];
ImgLinearUpdateVariableArrayWithId(head, i, id);
}
for(k=0; k<conjunct->nCluster; k++) {
tConjunct = conjunct->clusterArray[k];
supList = tConjunct->supList;
listSize = tConjunct->nSize;
for(j=0; j<listSize; j++) {
ImgLinearUpdateVariableArrayWithId(head, i, id);
}
}
}
bdd_free(bddOne);
head->bRefineVarArray = 0;
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearVariableArrayQuit | ( | Relation_t * | head | ) | [static] |
Function********************************************************************
Synopsis [Free variable array]
Description [Free variable array]
SideEffects []
SeeAlso []
Definition at line 4631 of file imgLinear.c.
{
int i;
VarLife_t **varArray;
varArray = head->varArray;
for(i=0; i<head->nVarArray; i++)
ImgLinearVariableLifeQuit(varArray[i]);
free(varArray);
head->varArray = 0;
head->nVarArray = 0;
memset(head->varArrayMap, -1, sizeof(int)*head->nVar);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearVariableLifeQuit | ( | VarLife_t * | var | ) | [static] |
Function********************************************************************
Synopsis [Free variable life array]
Description [Free variable lifearray]
SideEffects []
SeeAlso []
Definition at line 4660 of file imgLinear.c.
{
if(var->relArr) free(var->relArr);
free(var);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:| static void ImgLinearVariableOrder | ( | Relation_t * | head, |
| char * | baseName, | ||
| int | includeNS | ||
| ) | [static] |
Function********************************************************************
Synopsis [Make interface for variable ordering]
Description [Make interface for variable ordering]
SideEffects []
SeeAlso []
Definition at line 2727 of file imgLinear.c.
{
ImgLinearCAPOInterfaceVariableNodes(head, baseName, includeNS);
ImgLinearCAPOInterfaceVariableNet(head, baseName, includeNS);
ImgLinearCAPOInterfaceVariableScl(head, baseName);
ImgLinearCAPOInterfaceVariablePl(head, baseName, includeNS);
ImgLinearCAPOInterfaceAux(head, baseName);
ImgLinearCAPORun("MetaPlacer", baseName, 0);
ImgLinearCAPOReadVariableOrder(head, baseName, includeNS);
}
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:| static int linearCheckRange | ( | const void * | tc | ) | [static] |
Function********************************************************************
Synopsis [compare function for checking range of cluster]
Description [compare function for checking range of cluster]
SideEffects []
SeeAlso []
Definition at line 1973 of file imgLinear.c.
{
Cluster_t *tclu;
tclu = (Cluster_t *)tc;
if(tclu->from <= __clu->from && __clu->from <= tclu->to)
tclu->flag = 1;
if(tclu->from <= __clu->to && __clu->to <= tclu->to)
tclu->flag = 1;
return(1);
}
 Here is the caller graph for this function:
Here is the caller graph for this function:Variable Documentation
| Cluster_t* __clu |
Definition at line 62 of file imgLinear.c.
char linearVarString[] = "TCNI" [static] |
Definition at line 41 of file imgLinear.c.
char rcsid [] UNUSED = "$Id: imgLinear.c,v 1.18 2010/04/10 00:37:06 fabio Exp $" [static] |
CFile***********************************************************************
FileName [imgLinear.c]
PackageName [img]
Synopsis [Routines for image computation using Linear Arrangement published in TACAS02.]
Description []
SeeAlso []
Author [HoonSang Jin]
Copyright [Copyright (c) 1994-1996 The Regents of the Univ. of Colorado. All rights reserved.
Permission is hereby granted, without written agreement and without license or royalty fees, to use, copy, modify, and distribute this software and its documentation for any purpose, provided that the above copyright notice and the following two paragraphs appear in all copies of this software.
IN NO EVENT SHALL THE UNIVERSITY OF COLORADO BE LIABLE TO ANY PARTY FOR DIRECT, INDIRECT, SPECIAL, INCIDENTAL, OR CONSEQUENTIAL DAMAGES ARISING OUT OF THE USE OF THIS SOFTWARE AND ITS DOCUMENTATION, EVEN IF THE UNIVERSITY OF COLORADO HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
THE UNIVERSITY OF COLORADO SPECIFICALLY DISCLAIMS ANY WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. THE SOFTWARE PROVIDED HEREUNDER IS ON AN "AS IS" BASIS, AND THE UNIVERSITY OF COLORADO HAS NO OBLIGATION TO PROVIDE MAINTENANCE, SUPPORT, UPDATES, ENHANCEMENTS, OR MODIFICATIONS.]
Definition at line 40 of file imgLinear.c.