Mehran Pesteie

PhD Candidate, Robotics and Control Laboratory, Department of Electrical and Computer Engineering,
University of British Columbia.
ICICS X510, 2366 Main Mall, Vancouver, BC Canada V6T 1Z4
Email: mehranp[AT]ece.ubc.ca
Selected projects @UBC:
Adaptive augmentation of medical data using independently conditional variational auto-encoders
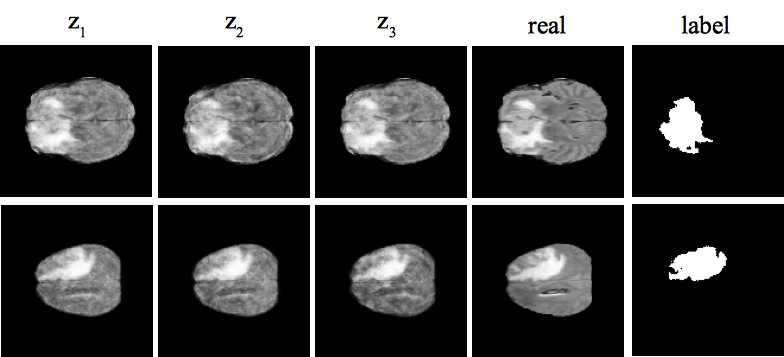
Despite their success in image recognition tasks, deep models require large amounts of data to be regularized. This is required in order to guarantee a minimum difference between train and test errors, which is crucial in clinical applications. However, small sample size is a typical characteristic of medical image datasets since clinical data collection is associated with high complexity and cost. On top of data acquisition, annotation and cleaning is often required prior to training, which adds a significant cost overhead. We aim to alleviate these limitations by proposing a variational generative model along with an effective data augmentation approach that utilizes the generative model to synthesize data to efficiently train a discriminative model. We demonstrate the effectiveness of the approach on two independent clinical datasets consisting of ultrasound images of the spine and magnetic resonance images of the brain.
[Paper] [AdaTrain Code] [ICVAE Code]
Deep Neural Maps for Unsupervised Visualization of High Grade Cancer in Prostate Biopsies
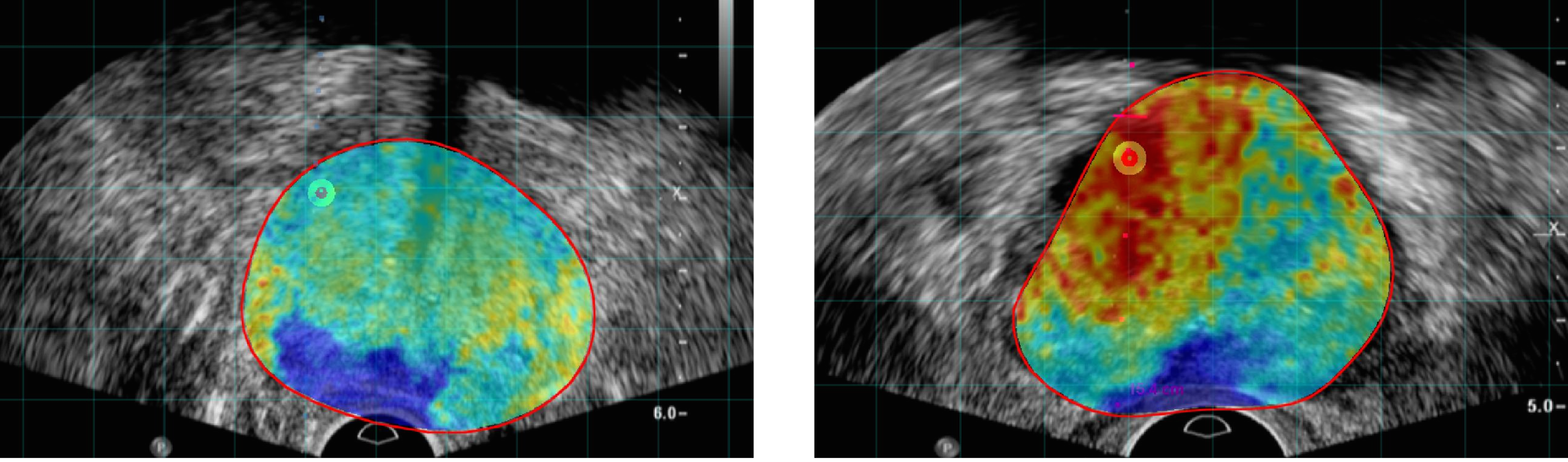
Prostate Cancer (PCa) is the most frequent noncutaneous cancer in men. Temporal Enhanced Ultrasound (TeUS), has emerged as a promising modality for PCa detection.
However, pathology labels are noisy and data from an entire core has a single label even when significantly heterogeneous.
Additionally, supervised methods are limited by data from cores with known pathology, and a significant portion of prostate data is discarded without being used.
We provide an end-to-end unsupervised solution to map PCa distribution from TeUS data using Deep Neural Maps (DNM).
TeUS data is transformed to a topologically arranged hyper-lattice, where similar samples are closer together in the lattice.
Therefore, similar regions of malignant and benign tissue in the prostate are clustered together.
Data from biopsy cores with known labels is used to associate the clusters with PCa.
Cancer probability maps generated using the unsupervised clustering of TeUS data helps intuitively visualize the distribution of abnormal tissue for augmenting TRUS-guided biopsies.
[Paper]
Deep Neural Maps

The deep neural map (DNM) is an unsupervised model that jointly learns an embedding of the input data and a mapping from the embedding space to an N-dimensional lattice. It consists of a convolutional auto-encoder (AE) and a self organizing map (SOM). The goal is to optimize the parameters of the AE and SOM such that for each data point, the l2 norm of its latent variable and its best matching unit in the SOM is minimized. Therefore, DNM provides a topographic map of the input where the locations of the neurons in the map indicate statistical features of the input patterns, such as category, style and pose. After training, the DNM can be used for data visualization, supervised and semi-supervised classification, and clustering.
Variational Mandible Shape Completion for Virtual Surgical Planning
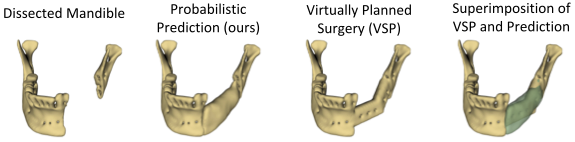
The premorbid geometry of the mandible is of significant relevance in jaw reconstructive surgeries and occasionally unknown to the surgical team. In this work, an optimization framework is introduced to train deep models for completion (reconstruction) of the missing segments of the bone based on the remaining healthy structure. To leverage the contextual information of the surroundings of the dissected region, the voxel-weighted Dice loss is introduced. To address the non-deterministic nature of the shape completion problem, we leverage a weighted multi-target probabilistic solution which is an extension to the conditional variational autoencoder (CVAE). This approach considers multiple targets as acceptable reconstructions, each weighted according to their conformity with the original shape.
[Paper] [Code]
Echocardiography Segmentation by Quality Translation Using Anatomically Constrained CycleGAN
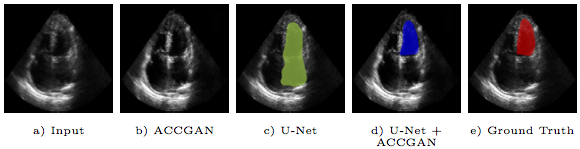
Segmentation of an echocardiogram (echo) is favorable for assessment of cardiac functionality and disease. The quality of the captured echo is a key factor that affects the segmentation accuracy. We propose a novel generative adversarial network architecture, which aims to improve echo quality for the segmentation of the left ventricle (LV). The proposed model is anatomically constrained to the structure of the LV in apical four chamber (AP4) echo view. A set of discriminative features are learned through unpaired translation of low to high quality echo using adversarial training. The anatomical constraint regularizes the model during end-to-end training to preserve the corresponding shape of the LV in the translated echo. Experiments show that leveraging information in the translated high quality echocardiograms by the proposed method improves the robustness of the segmentation.
[Paper]
Real-time plane classification of 3D ultrasound volumes of the lumbar spine.
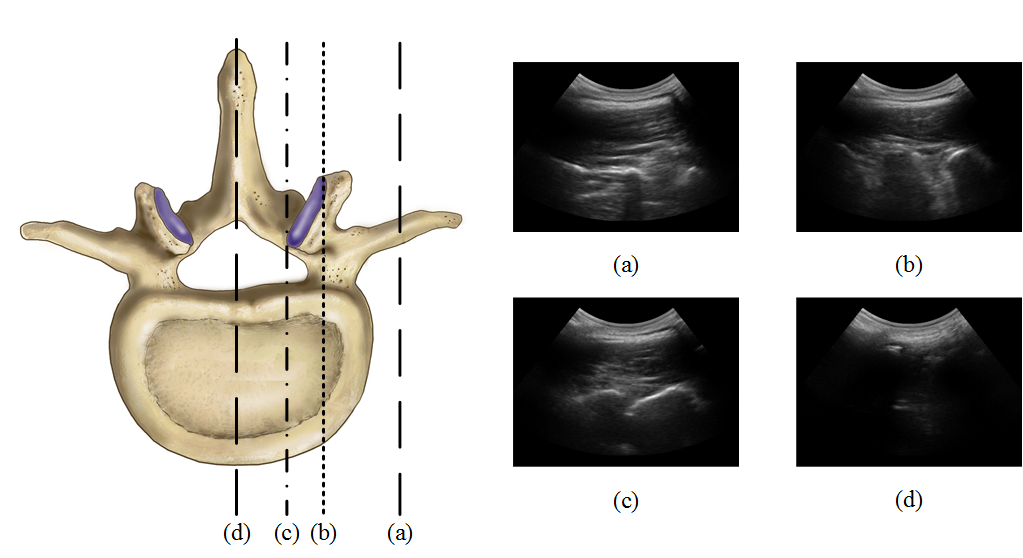
Injection therapy is a commonly used solution for back pain management. This procedure typically involves percutaneous insertion of a needle between or around the vertebrae, to deliver anesthetics near nerve bundles. Most frequently, spinal injections are either performed blindly using palpation, or under the guidance of fluoroscopy or Computed Tomography. Recently, due to the drawbacks of the ionizing radiation of such imaging modalities, there has been a growing interest in using ultrasound imaging as an alternative. However, the complex spinal anatomy with different wave-like structures (see the figure), affected by speckle noise, makes the accurate identification of the appropriate injection plane difficult. The aim of this study is to propose an automated system that can identify the optimal plane for epidural steroid injections and facet joint injections.
A hybrid deep network for automatic localization of the needle target in 3D ultrasound for spinal injections.
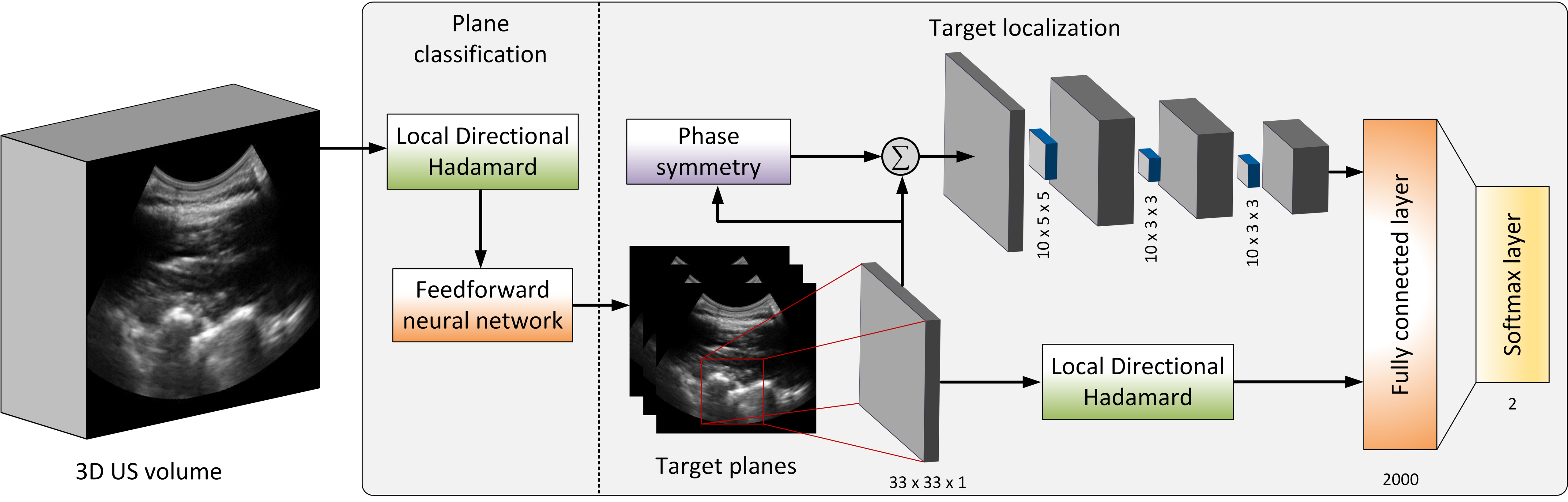
Accurate identification of the needle target is crucial for effective spinal injections. Currently, epidural needle placement is administered by a manual technique, relying on the sense of feel, which has a significant failure rate. Moreover, misleading the needle may lead to inadequate anesthesia, post dural puncture headaches and other potential complications. Ultrasound offers guidance to the physician for identification of the needle target, but accurate interpretation and localization remain challenges. We introduce a feature augmentation technique that incorporates convolutional feature maps, which are obtained by convolutional layers, with multi-scale local directional Hadamard features. Since the multi-scale Hadamard features are sensitive to the directionality of the US echoes from the surface of the vertebrae in the US image, the augmentation provides the deep network with a set of distinctive directional features from the sequency domain in addition to the feature maps automatically obtained from the spatial domain.
[Paper]
An open-source framework for deployment of deep learning models for image guided therapy.
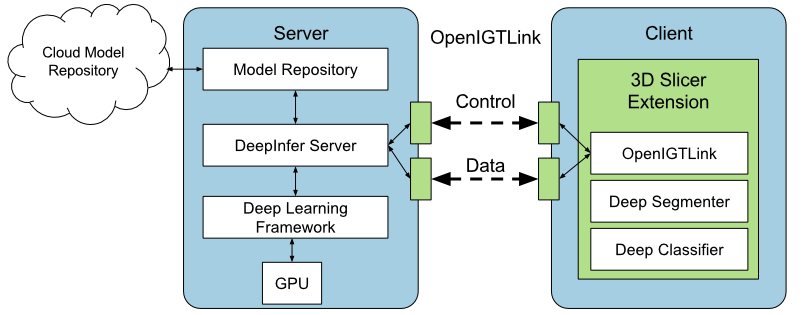
Deep learning models have outperformed some of the previous state-of-the-art approaches in medical image analysis. Instead of using hand-engineered features, deep models attempt to automatically extract hierarchical representations at multiple levels of abstraction from the data. However, utilizing deep models during image-guided therapy procedures requires integration of several software components which is often a tedious task for clinical researchers. Hence, there is a gap between the state-of-the-art machine learning research and its application in clinical setup. In this paper, we propose an open-source toolkit for medical image analysis with deep learning models in 3D Slicer, called "DeepInfer".
Unsupervised feature learning for tracking vertebrae in ultrasound.
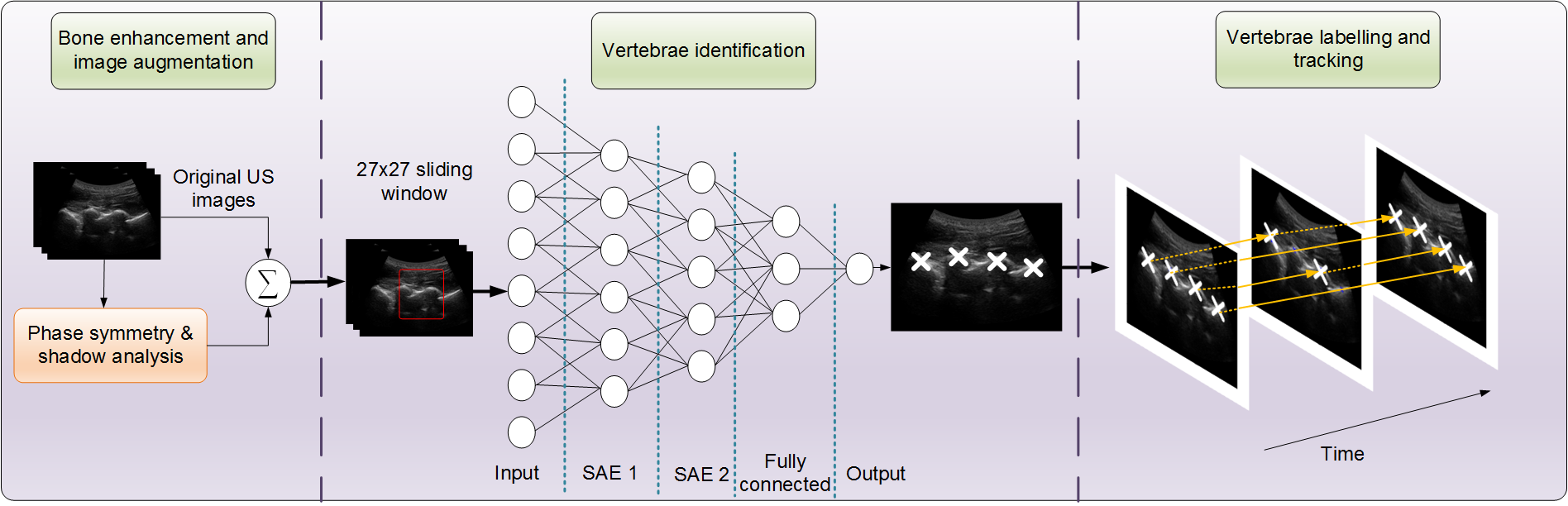
Percutaneous needle insertion procedures on the spine often require proper identification of the vertebral level in order to effectively deliver anesthetics and analgesic agents to achieve adequate block. In this study, we proposed a system to identify the vertebrae, assigning them to their respective levels, and track them in a standard sequence of ultrasound images, when imaged in the paramedian plane. In particular, a deep network is trained to automatically learn the anatomical features of the lamina peaks, and classify image patches, for pixel-level classification.
[Paper]